Digital Posters
Software Tools for Development, Data Processing & Analysis
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 15:00 - 16:00 |
3753.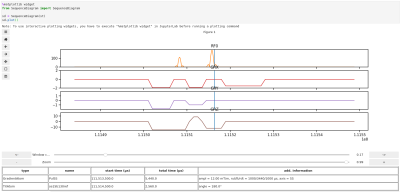 |
MRSequoia: A novel tool for MR sequence design, prototyping and validation.
Sebastian Hirsch1,2 and Stefan Hetzer1,2
1Berlin Center for Advanced Neuroimaging, Charité - Universitätsmedizin Berlin, Berlin, Germany, 2Bernstein Center for Computational Neuroscience, Berlin, Germany
MRSequoia is a novel open-source framework to aid MR sequence developers in the design, prototyping and validation of MR sequences. It can import the Siemens IDEA simulator output, and construct an abstract sequence timing from it, allowing for visualization of various aspects of the timing, as well as running custom or pre-defined timing checks. Furthermore, the timing can be exported to MRiLab for accurate spin-physics simulations, ensuring that identically the same timing is used in the simulation and on the scanner. MRSequoia is freely available and can easily be modified and extended thanks to its modular and object-oriented design.
|
|||
3754.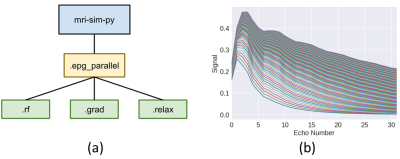 |
A GPU-accelerated Extended Phase Graph Algorithm for differentiable optimization and learning
Somnath Rakshit1, Ke Wang2, and Jonathan I Tamir3,4,5
1School of Information, The University of Texas at Austin, Austin, TX, United States, 2Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 3Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States, 4Diagnostic Medicine, Dell Medical School, The University of Texas at Austin, Austin, TX, United States, 5Oden Institute for Computational Engineering and Sciences, The University of Texas at Austin, Austin, TX, United States
The Extended Phase Graph Algorithm is a powerful tool for MRI sequence simulation and quantitative fitting, but such simulators are mostly written to run on CPU only and (with some exception) are poorly parallelized. A parallelized simulator compatible with other learning-based frameworks would be a useful tool to optimize scan parameters. Thus, we created an open source, GPU-accelerated EPG simulator in PyTorch. Since the simulator is fully differentiable by means of automatic differentiation, it can be used to take derivatives with respect to sequence parameters, e.g. flip angles, as well as tissue parameters, e.g. T1 and T2.
|
|||
3755.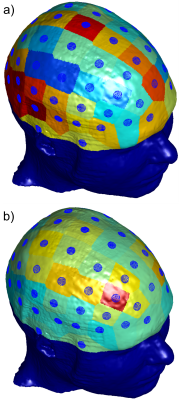 |
Investigation of TES Simulation Sensitivity to Skull Simplification using a Multimodal MR-Based Detailed Head Model
William Wartman1, Kyoko Fujimoto2, Mohammad Daneshzand3, Sergey Makarov1,3, and Aapo Nummenmaa3
1Electrical and Computer Engineering Department, Worcester Polytechnic Institute, Worcester, MA, United States, 2Center for Devices and Radiological Health, US Food and Drug Administration, Silver Spring, MD, United States, 3Athinoula A Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States
One of the most detailed multimodal human head models currently available (MIDA, 0.5 mm isotropic res., 100+ compartments) is accurately analyzed numerically to investigate the effects of the fine structure of the skull on transcranial electrical stimulation (TES). A simplified case, where the diploë and dura are treated as cortical bone, is compared against the full-model case. For each electrode in a 10-10 configuration, coefficients are computed independently to scale the cortical fields from the simplified case to match the cortical fields from the realistic case, investigating the topological field differences in the process.
|
|||
3756.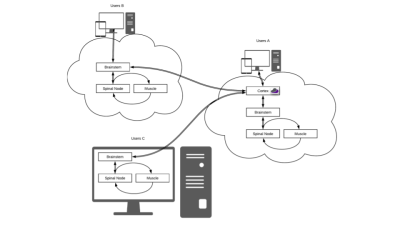 |
A web-accessible tool for rapid analytical simulations of MR coils via cloud computing
Eros Montin1,2, Giuseppe Carluccio1,2, and Riccardo Lattanzi1,2,3
1Department of Radiology, Bernard and Irene Schwartz Center for Biomedical Imaging, New York University School of Medicine, New York, NY, United States, 2Department of Radiology, Center for Advanced Imaging Innovation and Research (CAI2R), New York University School of Medicine, New York, NY, United States, 3Vilcek Institute of Graduate Biomedical Sciences, New York University School of Medicine, New York, NY, United States
DGF is a web-based application to simulate MR coils in the case of simple geometries that mimic actual anatomy. For example, spheres can model the head, whereas cylinders can model the torso, abdomen or extremities. Ultimate intrinsic performance limits can be calculated within the same framework and used as absolute references to evaluate coil designs. DGF relies on rapid analytical electrodynamic simulations based on dyadic Green’s functions, which are executed using Docker containers, either on local computers or via cloud computing. A web-GUI enables users to set up simulations and display results. DGF is part of the Cloud MR project.
|
|||
3757.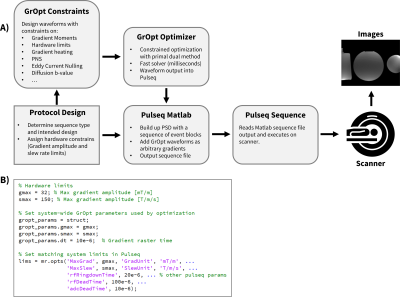 |
Using GrOpt with Pulseq for Easy Prototyping of Pulse Sequences with Optimized Waveforms
Michael Loecher1,2, Judith Zimmerman1,3, Matthew J Middione1,2, and Daniel B Ennis1,2,4
1Radiology, Stanford University, Stanford, CA, United States, 2Radiology, Veterans Affairs Health Care System, Palo Alto, CA, United States, 3Computer Science, Technical University of Munich, Garching, Germany, 4Cardiovascular Institute, Stanford University, Stanford, CA, United States
The objective of this work was to demonstrate the integration of two open-source MRI software packages: GrOpt and Pulseq. GrOpt allows for robust and fast optimization of gradient waveforms subject to various constraints, while Pulseq allows a flexible and straightforward approach to writing sequences and executing them on a scanner. We demonstrate example pulse sequences that integrate both software packages and show an imaging example from a PC-MRI experiment. The combination of GrOpt and Pulseq allows for optimized arbitrary gradient waveforms to be easily prototyped and run on a scanner.
|
|||
3758.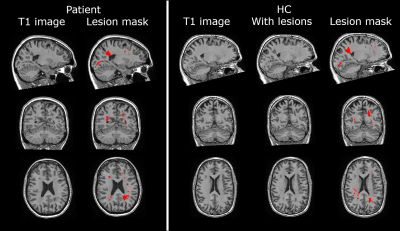 |
Lesion simulation software LESIM: a robust and flexible tool for realistic simulation of white matter lesions
Merlin M. Weeda1, Alexandra de Sitter1, Iman Brouwer1, Mitchell M. de Boer1, Rick J. van Tuijl1, Petra J.W. Pouwels1, Frederik Barkhof1,2, and Hugo Vrenken1
1Radiology and Nuclear Medicine, Amsterdam UMC - Location VUmc, Amsterdam, Netherlands, 2Institutes of Neurology and Healthcare Engineering UCL, London, United Kingdom
Multiple sclerosis(MS) is characterized by white matter(WM) lesions and grey matter(GM) atrophy in the central nervous system. Software to measure GM atrophy is severely hindered by the presence of lesions. In order to facilitate development of accurate GM segmentation software in the presence of WM lesions, we present a novel, robust, flexible and open-source lesion simulation tool: LESIM. Initial analysis with 25 LESIM lesion simulated images shows natural-looking lesions in the correct locations, with correct signal-to-noise ratio and intensity compared to the rest of the image. Analysis with FSL-SIENAX confirms that GM segmentation is affected in HCs with simulated lesions.
|
|||
3759.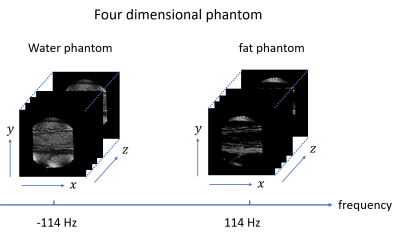 |
Creation of a four-dimensional numerical phantom for Bloch simulations of water-fat systems
Katsumi Kose1, Ryoichi Kose1, and Yasuhiko Terada2
1MRIsimulations Inc., Tokyo, Japan, 2University of Tsukuba, Tsukuba, Japan
A four-dimensional (4D) numerical phantom, which is defined by the three-dimensional (3D) spatial axes and the resonance frequency axis, is indispensable for Bloch simulations of protons in biological tissues with complex distribution of materials. In this study, a 4D phantom was created using an image dataset of an actual biological sample containing water and fat, and the Bloch simulation was performed using the phantom. As a result, 3D images of the samples containing water and fat were successfully reproduced, which demonstrated the usefulness of the concept of the proposed 4D phantom.
|
|||
3760. |
PyPulseq in a web browser: a zero footprint tool for collaborative and vendor-neutral pulse sequence development
Keerthi Sravan Ravi1,2, John Thomas Vaughan Jr.2, and Sairam Geethanath2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, New York, NY, United States
PyPulseq is a free, open-source and vendor-neutral pulse sequence development (PSD) tool, allowing users to develop MR sequences using the Python programming language. This work demonstrates running PyPulseq in a web browser. This is accomplished by leveraging Google Colab which enables executing arbitrary code in a web browser. A single-slice 2D Gradient Recalled Echo pulse sequence is programmed and executed on a 3T scanner and the acquired data is visualized. PyPulseq on Colab enables portable PSD. It is beneficial for educational purposes, collaborative PSD and for fostering reproducible acquisition methods.
|
|||
3761.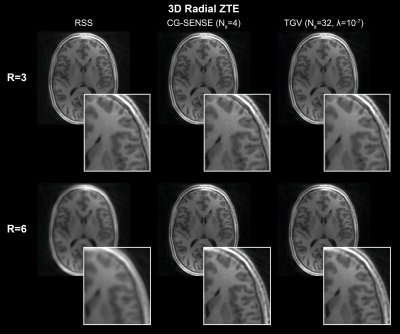 |
RIESLING: Radial Interstices Enable Speedy Low-volume Imaging
Tobias C Wood1, Emil Ljungberg1, and Florian Wiesinger2
1Neuroimaging, King's College London, London, United Kingdom, 2ASL Europe, GE Healthcare, Munich, Germany
We present an image reconstruction toolbox tuned for 3D radial ZTE images named Radial Interstices Enable Speedy Low-volume imagING (RIESLING). RIESLING matches the image quality of existing toolboxes while enabling fast reconstructions of high resolution ZTE datasets.
|
|||
3762.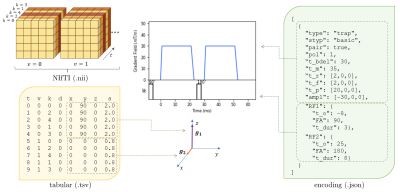 |
aDWI-BIDS: Advanced Diffusion Weighted Imaging Metadata for the Brain Imaging Data Structure
James Andrew Gholam1,2, Santiago Aja-Fernandez3, Matt Griffin1, Derek Jones2, Emre Kopanoglu2, Lars Mueller2, Markus Nilsson4, Filip Szczepankiewicz4, Chantal Tax2,5, Carl-Fredrik Westin6, and Leandro Beltrachini1,2
1School of Physics and Astronomy, Cardiff University, Cardiff, United Kingdom, 2CUBRIC, Cardiff University, Cardiff, United Kingdom, 3Universidad de Valladolid, Valladolid, Spain, 4Department of Diagnostic Radiology, Lund University, Lund, Sweden, 5Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 6Harvard Medical School, Boston, MA, United States
We present an extension to the Brain Imaging Data Structure (BIDS) to specialise it for diffusion weighted imaging. Detailed attribution of experimental parameters to regions of an aquisition is made possible with plain text files which remain compliant with BIDS. Complex diffusion encoding, slice-level diffusion encoding, and data collected with varying experimental parameters throughout the acquisition are all supported. Scope exists for reporting on RF pulses and gradient pulses in general within the sequence, without restriction to diffusion pulses.
|
|||
3763.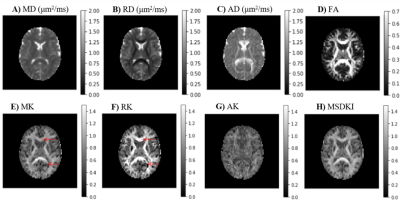 |
Diffusional Kurtosis Imaging in the Diffusion Imaging in Python Project
Rafael Neto Henriques1, Marta Correia2, Maurizio Marrale3, Elizabeth Huber4, John Kruper5, Serge Koudoro6, Jason Yeatman4,7, Eleftherios Garyfallidis6, and Ariel Rokem5
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Cognition and Brain Sciences Unit, University of Cambridge, Cambridge, United Kingdom, 3Department of Physics and Chemistry “Emilio Segrè”, University of Palermo, Palermo, Italy, 4Institute for Learning and Brain Science and Department of Speech and Hearing, University of Washington, Seattle, WA, United States, 5Department of Psychology and eScience Institute, The University of Washington, Seattle, WA, United States, 6Department of Intelligent Systems Engineering, Luddy School of Informatics, Computing and Engineering, Indiana University Bloomington, Bloomington, IN, United States, 7Department of Pediatrics and Graduate School of Education, Stanford University, Stanford, CA, United States
Diffusion Kurtosis Imaging (DKI) estimates non-Gaussian diffusion in biological tissue from diffusion-weighted MRI, providing a useful marker for individual differences in tissue microstructure. We present a well-tested, well-documented open-source implementation of DKI as part of the DIPY (Diffusion Imaging in Python) project. The implementation provides standard DKI metrics, as well as extensions of the method for microstructure modeling and tractography. We demonstrate the use of these methods in openly available datasets.
|
|||
3764.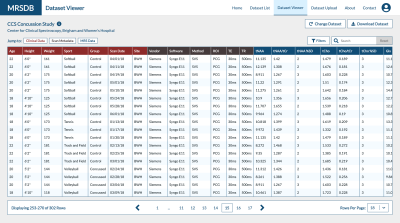 |
MRSDB: A Scalable Multisite Data Library for Clinical and Machine Learning Applications of Magnetic Resonance Spectroscopy
Sam H. Jiang1, Eduardo Coello1, Marcia S. Louis1, Katherine M. Breedlove1, and Alexander P. Lin1
1Center for Clinical Spectroscopy, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, United States
This work introduces a mock standardized library of multisite magnetic resonance spectroscopy (MRS) data and functional online application called MRS Database (MRSDB) for the secure collection, processing, and sharing of that data. The goal of this platform is to enhance global collaboration, improve objective diagnostics in radiology, and facilitate the development of machine learning and artificial intelligence techniques using MRS.
|
|||
3765.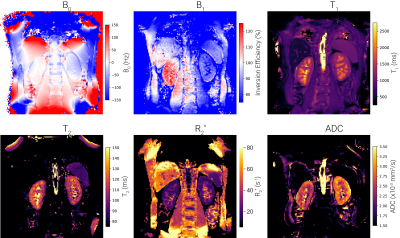 |
UKRIN Kidney Analysis Toolbox (UKAT): A Framework for Harmonized Quantitative Renal MRI Analysis
Alexander J Daniel1, Fabio Nery2, João Sousa3, Charlotte E Buchanan1, Hao Li4, Andrew N Priest4,5, Steven Sourbron3, David L Thomas6,7,8, and Susan T Francis1
1Sir Peter Mansfield Imaging Centre, University of Nottingham, Nottingham, United Kingdom, 2Great Ormond Street Institute of Child Health, University College London, London, United Kingdom, 3Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 4Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 5Department of Radiology, Addenbrooke’s Hospital, Cambridge, United Kingdom, 6Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 7Dementia Research Centre, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 8Wellcome Centre for Human Neuroimaging, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom
Multicentre validation studies are key to the clinical translation of renal MRI and as such, the development of harmonised, cross vendor protocols is crucial. To process data acquired from these protocols, the UK Renal Imaging Network Kidney Analysis Toolbox (UKAT) has been developed. This open-source, vendor agnostic and easy to use Python package can be used for image registration, field mapping, relaxometry and diffusion mapping. UKATs combination of robust software, documented methodological decisions and easy to follow tutorials means we envisage this as a useful tool for the renal and abdominal imaging community.
|
|||
3766.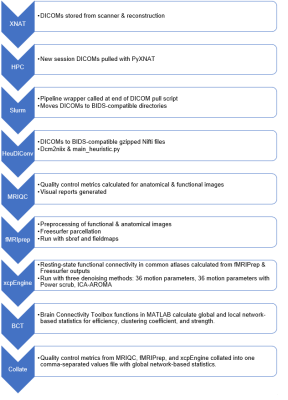 |
Automating Reproducible Connectivity Processing Pipelines on High Performance Computing Machines
Paul B Camacho1,2,3, Evan D Anderson3,4, Aaron T Anderson3,5, Hillary Schwarb3, Tracey M Wszalek3, and Brad P Sutton1,2,3
1Neuroscience Program, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Decision Neuroscience Laboratory, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 5Stephens Family Clinical Research Institute, Carle Foundation Hospital, Urbana, IL, United States
Automating pipelines open-source reproducible toolkit Docker containers for data quality control, processing, and analysis maximizes data value and minimizes time spent performing manual tests. Achieving high throughput with these pipelines requires more computational resources than a standard laboratory workstation, leading to migrating pipelines to high-performance computing systems. We created an open-source wrapper for higher security Singularity images required for resting-state functional connectivity workflows on high-performance computing systems, which extends function to report collection, network-based statistics, and versioning documentation. This pipeline was then tested with an existing aging and cognition data set for benchmarking and demonstration.
|
|||
3767.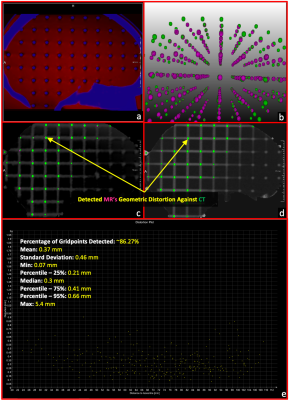 |
Development of an online distortion measurement prototype
Lumeng Cui1, Johanna Grigo2, Gerald R. Moran3, and Niranjan Venugopal4
1Division of Biomedical Engineering, University of Saskatchewan, Saskatoon, SK, Canada, 2Universitätsklinikum Erlangen, Erlangen, Germany, 3Research Collaboration Manager, Siemens Healthcare Limited, Oakville, ON, Canada, 4Department of Radiology, University of Manitoba, Winnipeg, MB, Canada
Magnetic Resonance Imaging (MRI) is increasingly essential in radiation therapy (RT) planning and so does the measurement of geometric distortion latent in MR imaging. In this work, we have developed a prototype that can accommodate any type of grid-like phantoms and provides an on-the-scanner software solution for the analysis of spatial distortion MR images. The prototype takes a prior CT scan and a new acquired MR dataset as inputs and generates a qualitative visualization and quantitative evaluation for the spatial distortion in the MRI volume. The prototype was assessed with two grid-like phantoms with good success.
|
|||
3768.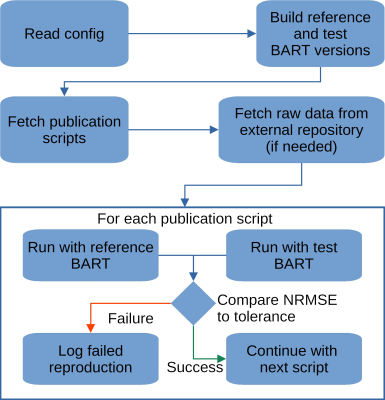 |
Reproducibility meets Software Testing: Automatic Tests of Reproducible Publications Using BART
H. Christian M. Holme1,2 and Martin Uecker1,2,3
1Institute for Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2Partner Site Göttingen, DZHK (German Centre for Cardiovascular Research), Göttingen, Germany, 3Campus Institute Data Science (CIDAS), University of Göttingen, Göttingen, Germany
Progress in science is only possible if we can trust our current knowledge and build on it. Thus, it is not only important to ensure reproducibility of published results but also to make sure that this is achieved in such a way that building on it is possible. Therefore, we describe a workflow and tool to verify the reproducibility of publications that make use of the BART toolbox. This ensures that published results remain reproducible into the future, even using newer versions of BART.
|
|||
3769.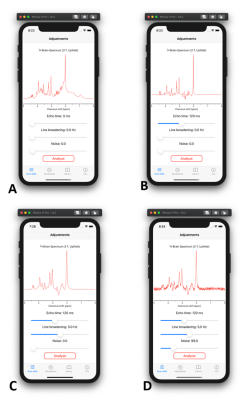 |
Mobile application for in vivo MR spectroscopy: Pocket MRS
Martin Gajdošík1, Karl Landheer1, and Christoph Juchem1,2
1Department of Biomedical Engineering, Columbia University, New York City, NY, United States, 2Department of Radiology, Columbia University Medical Center, New York, NY, United States
Pocket MRS is a mobile application that offers simple and easy access to simulated spectra from human brain and its detailed analysis. Spectra available from 0 to 300 ms TE were simulated using realistic quantum mechanical density operator simulations, and scaled using known concentration, T1 and T2 values across 19 different metabolites and a sum of 10 macromolecules. Spectra can be manipulated with respect to echo time, magnetic field quality and noise levels, providing quick and convenient visualization of the impact of typical experimental conditions on spectral appearance.
|
|||
3770.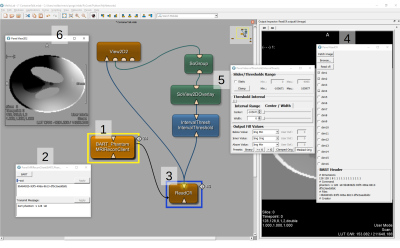 |
Visual Remote Control of MRI Reconstruction Toolboxes
Robin Niklas Wilke1, Simon Konstandin1, Daniel Christopher Hoinkiss1, Martin Uecker2,3, and Matthias Günther1,4
1Fraunhofer Institute for Digital Medicine MEVIS, Bremen, Germany, 2DZHK (German Centre for Cardiovascular Research), Partner Site Goettingen, Berlin, Germany, Goettingen, Germany, 3Institute for Diagnostic and Interventional Radiology, University Medical Center Goettingen, Goettingen, Germany, 4MR-Imaging and Spectroscopy, Faculty 01 (Physics/Electrical Engineering), University of Bremen, Bremen, Germany
Commercial MRI software that is shipped with an MRI scanner is typically not suited for research and development in state-of-the-art MRI imaging schemes. In particular, the advanced MR imaging requires both high computing power and dedicated frameworks for (iterative) algorithms. Reconstruction frameworks tend to be rather optimized for MR reconstruction but usability, rapid prototyping and medical image analyses. We overcome this limit by providing a module interface in a medical image processing and visualization toolbox that enables remote control of virtualized MRI reconstruction toolboxes with an interface to the output data.
|
|||
3771.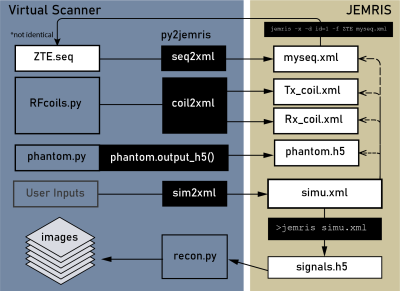 |
Bridging Open Source Sequence Simulation and Acquisition with py2jemris
Gehua Tong1, Sairam Geethanath2, and John Thomas Vaughan, Jr.2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center, Columbia University, New York, NY, United States
Open-source sequence development tools are often simulation-focused or acquisition-focused. The ability to simulate and acquire from the same sequence file would help speed up method development. In this work, we present an open-source Python tool, py2jemris, which converts arbitrary Pulseq files to JEMRIS simulation format with a zero-footprint Google Colab notebook. A dual simulation/acquisition experiment using the same sequence file was performed to demonstrate the development pipeline. The conversion and simulation time were recorded and evaluated.
|
|||
3772.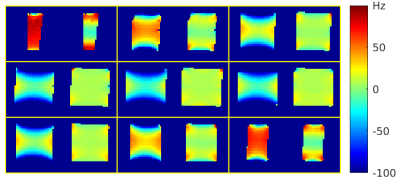 |
An open toolbox for harmonized B0 shimming
Jon-Fredrik Nielsen1, Berkin Bilgic2,3, Jason P Stockmann2, Borjan Gagoski4, Jr-Yuan George Chiou5, Lipeng Ning6, Yang Ji6, Yogesh Rathi5, Jeffrey A Fessler7, Douglas C Noll1, and Maxim Zaitsev8
1fMRI Laboratory, University of Michigan, Ann Arbor, MI, United States, 2Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 3Harvard Medical School, Boston, MA, United States, 4Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 5Radiology, Brigham and Women’s Hospital, Boston, MA, United States, 6Psychiatry, Brigham and Women’s Hospital, Boston, MA, United States, 7Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 8High Field MR Center, Center for Medical Physics and Biomedical Engineering, Medical University of Vienna, Vienna, Austria
Commercial MRI scanners are typically equipped with linear and 2nd-order (spherical harmonic) shim channels that offer some control of the B0 field, but the shim settings are usually adjusted automatically and non-transparently during the scanner’s prescan routine with little or no user input. This practice makes it difficult to ensure consistent experimental conditions across sessions and sites, and may lead to suboptimal shim settings for a given application. We introduce an open toolbox for ‘harmonized’ B0 shimming across sites and vendor platforms, that makes full use of all available shim channels.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.