Digital Posters
Machine Learning: Multimodal & Multireader Insights
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 19:00 - 20:00 |
2417.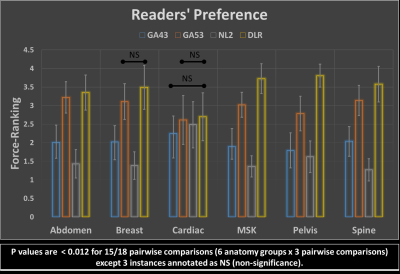 |
Prospective Performance Evaluation of the Deep Learning Reconstruction Method at 1.5T: A Multi-Anatomy and Multi-Reader Study
Hung Do1, Mo Kadbi1, Dawn Berkeley1, Brian Tymkiw1, and Erin Kelly1
1Canon Medical Systems USA, Inc., Tustin, CA, United States
We prospectively evaluate the generalized performance of the Deep Learning Reconstruction (DLR) method on 55 datasets acquired from 16 different anatomies. For each pulse sequence in each of the 16 anatomies, DLR and 3 predicate methods were reconstructed for randomized blinded review by 3 radiologists based on 8 scoring criteria plus a force-ranking. DLR was scored statistically higher than all 3 predicate methods in 92% of the pairwise comparisons in terms of overall image quality, clinically relevant anatomical/pathological features, and force-ranking. This work demonstrates that DLR generalizes to various anatomies and is frequently preferred over existing methods by experienced readers.
|
|||
2418.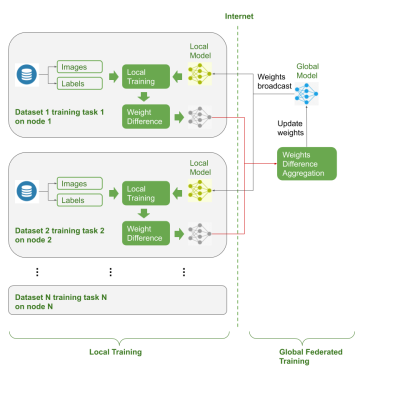 |
Federated Multi-task Image Classification on Heterogeneous Medical data with Privacy Perversing
Shenjun Zhong1, Adam Morris2, Zhaolin Chen1, and Gary Egan1
1Monash Biomedical Imaging, Monash University, Australia, Melbourne, Australia, 2Monash eResearch Center, Monash University, Australia, Melbourne, Australia
There is a lack of pre-trained deep learning model weights on large scale medical image dataset, due to privacy concerns. Federated learning enables training deep networks while preserving privacy. This work explored co-training multi-task models on multiple heterogeneous datasets, and validated the usage of federated learning could serve the purpose of pre-trained weights for downstream tasks.
|
|||
2419.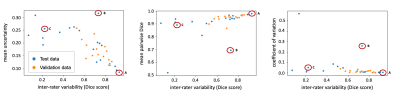 |
Do you Agree? An Exploration of Inter-rater Variability and Deep Learning Segmentation Uncertainty
Katharina Viktoria Hoebel1,2, Christopher P Bridge1,3, Jay Biren Patel1,2, Ken Chang1,2, Marco C Pinho1, Xiaoyue Ma4, Bruce R Rosen1, Tracy T Batchelor5, Elizabeth R Gerstner1,5, and Jayashree Kalpathy-Cramer1
1Athinoula A. Martinos Center for Biomedical Imaging, Boston, MA, United States, 2Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA, United States, 3MGH and BWH Center for Clinical Data Science, Boston, MA, United States, 4Department of Magnetic Resonance, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 5Stephen E. and Catherine Pappas Center for Neuro-Oncology, Massachusetts General Hospital, Boston, MA, United States
The outlines of target structures on medical imaging can be highly ambiguous. The uncertainty about the “true” outline is evident in high inter-rater variability of manual segmentations. So far, no method is available to identify cases likely to exhibit a high inter-rater variability. Here, we demonstrate that ground truth independent uncertainty metrics extracted from a MC dropout segmentation model developed on labels of only one rater correlate with inter-rater variability. This relationship can be used to identify ambiguous cases and flag them for more detailed review supporting consistent and reliable patient evaluation in research and clinical settings.
|
|||
2420.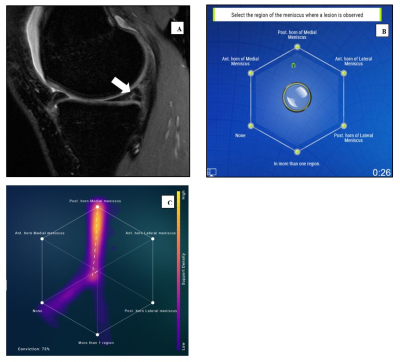 |
Swarm intelligence: a novel clinical strategy for improving imaging annotation accuracy, using wisdom of the crowds.
Rutwik Shah1, Bruno Astuto Arouche Nunes1, Tyler Gleason1, Justin Banaga1, Kevin Sweetwood1, Allen Ye1, Will Fletcher1, Rina Patel1, Kevin McGill1, Thomas Link1, Valentina Pedoia1, Sharmila Majumdar1, and Jason Crane1
1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States
Radiologists play a central role in image annotation for training Machine Learning models. Key challenges in this regard include low inter-reader agreement for challenging cases and concerns of interpersonal bias amongst trainers. Inspired by biological swarm intelligence, we explored the use of real time consensus labeling by three sub-specialty (MSK) trained radiologists and five radiology residents in improving training data. A second swarm session with three residents was conducted to explore the effect of swarm size. These results were validated against clinical ground truth and also compared with results from a state-of-the-art AI model tested on the same dataset.
|
|||
2421.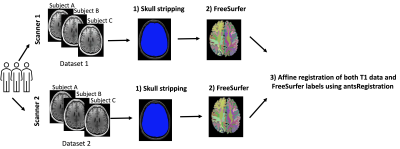 |
Harmonization of multi-site T1 data using CycleGAN with segmentation loss (CycleGANs)
Suheyla Cetin-Karayumak1, Evdokiya Knyazhanskaya2, Brynn Vessey2, Sylvain Bouix1, Benjamin Wade3, David Tate4, Paul Sherman5, and Yogesh Rathi1
1Brigham and Women's Hospital and Harvard Medical School, Boston, MA, United States, 2Brigham and Women's Hospital, Boston, MA, United States, 3Ahmanson-Lovelace Brain Mapping Center, UCLA, Los Angeles, CA, United States, 4University of Utah, Salt Lake City, UT, United States, 5U.S. Air Force School of Aerospace Medicine, San Antonio, TX, United States
This study aims to tackle the structural MRI (T1) data harmonization problem by presenting a novel multi-site T1 data harmonization, which uses the CycleGAN network with segmentation loss (CycleGANs). CycleGANs aims to learn an efficient mapping of T1 data across scanners from the same set of subjects while simultaneously learning the mapping of free surfer parcellations. We demonstrated the efficacy of the method with the Dice overlap scores between FreeSurfer parcellations across two datasets before and after harmonization.
|
|||
2422.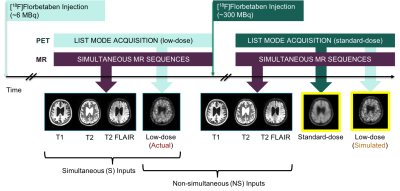 |
Does Simultaneous Morphological Inputs Matter for Deep Learning Enhancement of Ultra-low Amyloid PET/MRI?
Kevin T. Chen1, Olalekan Adeyeri2, Tyler N Toueg3, Elizabeth Mormino3, Mehdi Khalighi1, and Greg Zaharchuk1
1Radiology, Stanford University, Stanford, CA, United States, 2Salem State University, Salem, MA, United States, 3Neurology and Neurological Sciences, Stanford University, Stanford, CA, United States
We have previously generated diagnostic quality amyloid positron emission tomography (PET) images with deep learning enhancement of actual ultra-low-dose (~2% of the original) PET images and simultaneously acquired structural magnetic resonance imaging (MRI) inputs. Here, we will investigate whether simultaneity is a requirement for such structural MRI inputs. If simultaneity is not required, this will increase the utility of MRI-assisted ultra-low-dose PET imaging by including data acquired on separate PET/ computed tomography (CT) and standalone MRI machines.
|
|||
2423.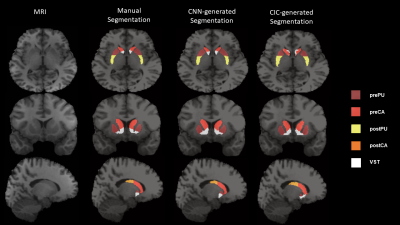 |
Multi-Task Learning based 3-Dimensional Striatal Segmentation of MRI – PET and fMRI Objective Assessment
Mario Serrano-Sosa1, Jared Van Snellenberg2, Jiayan Meng2, Jacob Luceno2, Karl Spuhler3, Jodi Weinstein2, Anissa Abi-Dargham2, Mark Slifstein2, and Chuan Huang2,4
1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Psychiatry, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States, 3Radiation Oncology, NYU Langone, New York, NY, United States, 4Radiology, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States
Segmenting striatal subregions can be difficult; wherein atlas-based approaches have been shown to be less reliable in patient populations and have problems segmenting smaller striatal ROI’s. We developed a Multi-Task Learning model to segment multiple 3D striatal subregions using a Convolutional Neural Network and compared it to the Clinical Imaging Center atlas (CIC). Dice Score Coefficient and multi-modal objective assessment (PET and fMRI) were conducted to evaluate the reliability of MTL-generated segmentations compared to atlas-based. Overall, MTL-generated segmentations were more comparable to manual than CIC across all ROI’s and analyses. Thus, we show MTL method provides reliable striatal subregion segmentations.
|
|||
2424.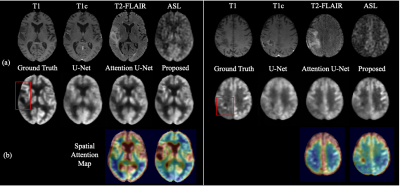 |
Zero-dose FDG PET Brain Imaging
Jiahong Ouyang1, kevin Chen2, and Greg Zaharchuk2
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States
PET is a widely used imaging technique but it requires exposing subjects to radiation and is not offered in the majority of medical centers in the world. Here, we proposed to synthesize FDG-PET images from multi-contrast MR images by a U-Net based network with symmetry-aware spatial-wise attention, channel-wise attention, split-input modules, and random dropout training strategy. The experiments on a brain tumor dataset of 70 patients demonstrated that the proposed method was able to generate high-quality PET from MR images without the need for radiotracer injection. We also demonstrate methods to handle potential missing or corrupted sequences.
|
|||
2425.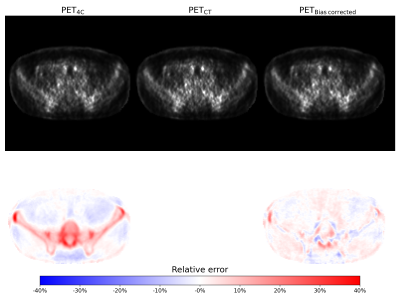 |
Bias correction for PET/MR attenuation correction using generative adversarial networks
Bendik Skarre Abrahamsen1, Tone Frost Bathen1,2, Live Eikenes1, and Mattijs Elschot1,2
1Department of Circulation and Medical Imaging, NTNU, Trondheim, Norway, 2Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
Attenuation correction is a challenge in PET/MRI. In this study we propose a novel attenuation correction method based on estimating the bias image between PET reconstructed using a 4-class attenuation correction map and PET reconstructed with an attenuation correction map where bone information is added from a co-registered CT image. A generative adversarial network was trained to estimate the bias between the PET images. The proposed method has comparable performance to other Deep Learning based attenuation correction methods where no additional MRI sequences are acquired. Bias estimation thus constitutes a viable alternative to pseudo-CT generation for PET/MR attenuation correction.
|
|||
2426.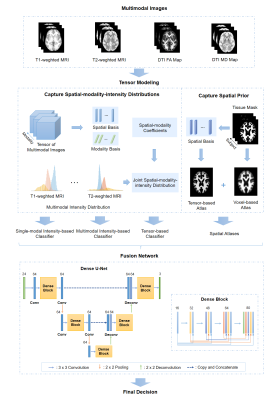 |
Multimodal Image Fusion Integrating Tensor Modeling and Deep Learning
Wenli Li1, Ziyu Meng1, Ruihao Liu1, Zhi-Pei Liang2,3, and Yao Li1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Multimodal brain imaging acquires complementary information of the brain. However, due to the high dimensionality of the data, it is challenging to capture the underlying joint spatial and cross-modal dependence required for statistical inference in various brain image processing tasks. In this work, we proposed a new multimodal image fusion method that synergistically integrates tensor modeling and deep learning. The tensor model was used to capture the joint spatial-intensity-modality dependence and deep learning was used to fuse spatial-intensity-modality information. Our method has been applied to multimodal brain image segmentation, producing significantly improved results.
|
|||
2427.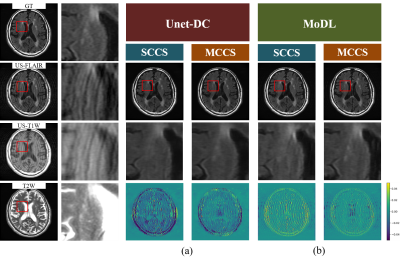 |
Multi-contrast CS reconstruction using data-driven and model-based deep neural networks
Tomoki Miyasaka1, Satoshi Funayama2, Daiki Tamada2, Utaroh Motosugi3, Hiroyuki Morisaka2, Hiroshi Onishi2, and Yasuhiko Terada1
1Institute of Applied Physics, University of Tsukuba, Tsukuba, Japan, 2Department of Radiology, University of Yamanashi, Chuo, Japan, 3Department of Radiology, Kofu-Kyoritsu Hospital, Kofu, Japan
The use of deep learning (DL) for compressed sensing (CS) have recently received increased attention. Generally, DL-CS uses single-contrast CS reconstruction (SCCS) where the single-contrast image is used as the network input. However, in clinical routine examinations, different contrast images are acquired in the same session, and CS reconstruction using multi-contrast images as the input (MCCS) has the potential to show better performance. Here, we applied DL-MCCS to brain MRI images acquired during routine examinations. We trained data-driven and model-based networks, and showed that for both cases, MCCS outperformed SCCS.
|
|||
2428.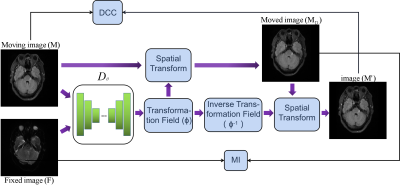 |
Unsupervised deep learning for multi-modal MR image registration with topology-preserving dual consistency constraint
Yu Zhang1, Weijian Huang1, Fei Li1, Qiang He2, Haoyun Liang1, Xin Liu1, Hairong Zheng1, and Shanshan Wang1
1Paul C Lauterbur Research Center, Shenzhen Inst. of Advanced Technology, shenzhen, China, 2United Imaging Research Institute of Innovative Medical Equipment, Shenzhen, China
Multi-modal magnetic resonance (MR) image registration is essential in the clinic to achieve accurate imaging-based disease diagnosis and treatment planning. Although the existing registration methods have achieved good performance and attracted widespread attention, the image details may be lost after registration. In this study, we propose a multi-modal MR image registration with topology-preserving dual consistency constraint, which achieves the best registration performance with a Dice score of 0.813 in identifying stroke lesions.
|
|||
2429.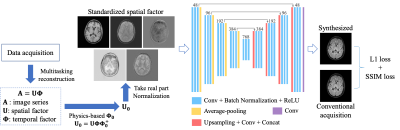 |
Direct Synthesis of Multi-Contrast Images from MR Multitasking Spatial Factors Using Deep Learning
Shihan Qiu1,2, Yuhua Chen1,2, Sen Ma1, Zhaoyang Fan1,2, Anthony G. Christodoulou1,2, Yibin Xie1, and Debiao Li1,2
1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 2Department of Bioengineering, UCLA, Los Angeles, CA, United States
MR Multitasking is an efficient approach for quantification of multiple parametric maps in a single scan. The Bloch equations can be used to derive conventional contrast-weighted images, which are still preferred by clinicians for diagnosis, from quantitative maps. However, due to imperfect modeling and acquisition, these synthetic images often exhibit artifacts. In this study, we developed a deep learning-based method to synthesize contrast-weighted images from Multitasking spatial factors without explicit Bloch modeling. We demonstrated that our method provided synthetic images with higher quality and fidelity than the model-based approach or a similar deep learning method using quantitative maps as input.
|
|||
2430.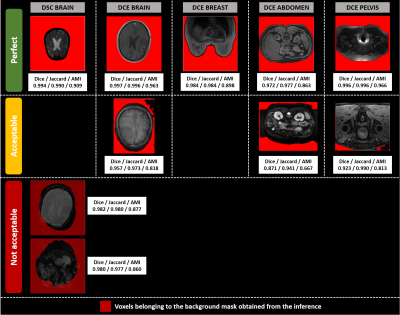 |
Multi-sequence and multi-regional background segmentation on multi-centric DSC and DCE MRI using deep learning
Henitsoa RASOANANDRIANINA1, Anais BERNARD1, Guillaume GAUTIER1, Julien ROUYER1, Yves HAXAIRE2, Christophe AVARE3, and Lucile BRUN1
1Department of Research & Innovation, Olea Medical, La Ciotat, France, 2Clinical Program Department, Olea Medical, La Ciotat, France, 3Avicenna.ai, La Ciotat, France
In this study, we present an automatic, multi-regional and multi-sequence deep-learning-based algorithm for background segmentation on both DSC and DCE images which consisted of a 2D U-net trained with a large multi-centric and multi-vendor database including DSC brain, DCE brain, DCE breast, DCE abdomen and DCE pelvis data. Cross-validation-based training results showed an overall good performance of the proposed algorithm with a median Dice score of 0.974 in test set and 0.979 over all datasets, and a median inference duration of 0.15s per volume on GPU. This is the first reported deep-learning-based multi-sequence and multi-regional background segmentation on MRI data.
|
|||
2431.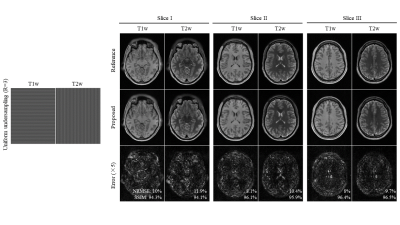 |
Multi-Contrast MRI Reconstruction from Single-Channel Uniformly Undersampled Data via Deep Learning
Christopher Man1,2, Linfang Xiao1,2, Yilong Liu1,2, Vick Lau1,2, Zheyuan Yi1,2,3, Alex T. L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
This study presents a deep learning based reconstruction for multi-contrast MR data with orthogonal undersampling directions across different contrasts. It enables exploiting the rich structural similarities from multiple contrasts as well as the incoherency arose from complementary sampling. The results show that the proposed method can achieve robust reconstruction for single-channel multi-contrast MR data at R=4.
|
|||
2432.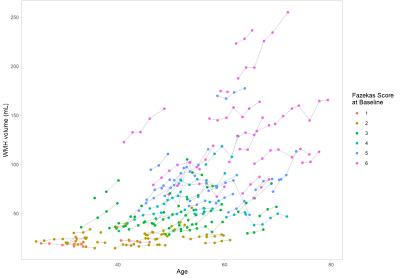 |
Automated assessment of longitudinal White Matter Hyperintensities changes using a novel convolutional neural network in CADASIL
Valentin Demeusy1, Florent Roche1, Fabrice Vincent1, Jean-Pierre Guichard2, Jessica Lebenberg3,4, Eric Jouvent3,5, and Hugues Chabriat3,5
1Imaging Core Lab, Medpace, Lyon, France, 2Department of Neuroradiology, Hôpital Lariboisière, APHP, Paris, France, 3FHU NeuroVasc, INSERM U1141, Paris, France, 4Université de Paris, Paris, France, 5Departement of Neurology, Hôpital Lariboisière, APHP, Paris, France
We propose a novel automatic WMH segmentation method based on a convolutional neural network to study the longitudinal WMH changes among a cohort of 101 CADASIL patients. We demonstrate that this method is able to produce consistent quantitative measures of WMH volume by the strong correlation between the computed baseline WMH volume and the clinically assessed Fazekas score. Our main results show that the progression of WMH is correlated to the baseline volume and that this progression largely vary at individual level although a rapid extension is mainly detected between 40 and 60 years in the whole population.
|
|||
2433.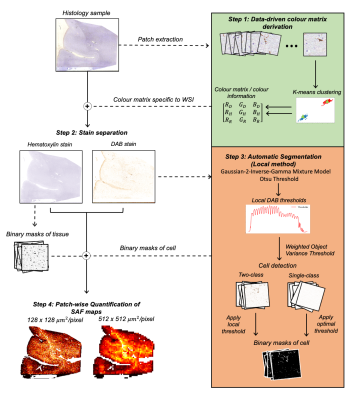 |
Automatic extraction of reproducible semi-quantitative histological metrics for MRI-histology correlations
Daniel ZL Kor1, Saad Jbabdi1, Jeroen Mollink1, Istvan N Huszar1, Menuka Pallebage- Gamarallage2, Adele Smart2, Connor Scott2, Olaf Ansorge2, Amy FD Howard1, and Karla L Miller1
1Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 2Neuropathology, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
Immunohistochemistry (IHC) images are often used as a microscopic validation tool for MRI. Acquisition of MRI and IHC in the same ex-vivo tissue sample can enable direct correlation between MRI measures and purported sources of image contrast derived from IHC, ideally at the voxel level. However, most IHC analyses still involve manual intervention (e.g. setting of thresholds). Here, we describe an end-to-end pipeline for automatically extracting stained area fraction maps to quantify the IHC stain for a given microstructural feature. The pipeline has improved reproducibility and robustness to histology artefacts, compared to manual MRI-histology analyses that suffer from inter-operator bias.
|
|||
2434.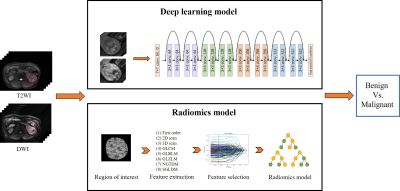 |
MRI-based deep learning model in differentiating benign from malignant renal tumors: a comparison study with radiomics analysis
Qing Xu1, Weiqiang Dou2, and Jing Ye1
1Northern Jiangsu People's Hospital, Yangzhou, China, 2GE Healthcare, MR Research China, Beijing, China
The aim of this study was to evaluate the feasibility of magnetic resonance imaging (MRI) based deep learning (DL) model in differentiating benign from malignant renal tumors. The performance of the applied DL model was further compared with that from a random forest radiomics model. More robust performance was achieved using MRI based DL model than the radiomics model (AUC = 0.925 vs 0.854, p<0.05). Therefore, the applied MRI based deep transfer learning model might be considered a convenient and reliable approach for differentiating benign from malignant renal tumors in clinic.
|
|||
2435.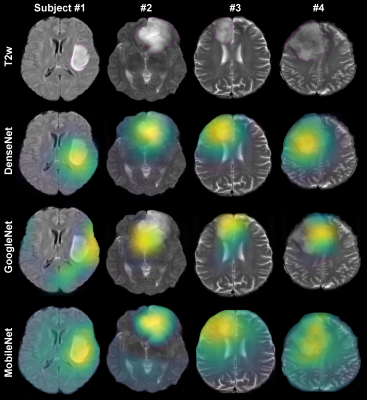 |
Evaluation of Automated Brain Tumor Localization by Explainable Deep Learning Methods
Morteza Esmaeili1, Vegard Antun2, Riyas Vettukattil3, Hassan Banitalebi1, Nina Krogh1, and Jonn Terje Geitung1,3
1Akershus University Hospital, Lørenskog, Norway, 2Department of Mathematics, University of Oslo, Oslo, Norway, 3Faculty of Medicine, Institute of Clinical Medicine, University of Oslo, Oslo, Norway
Machine learning approaches provide convenient autonomous object classification in medical imaging domains. This study uses an explainable method to evaluate the high-level features of deep learning methods in tumor localization.
|
|||
2436.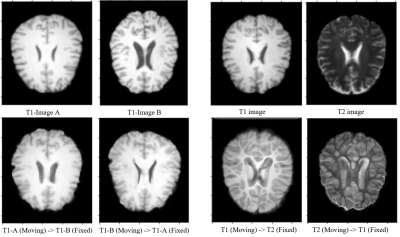 |
A Comparative Study of Deep Learning Based Deformable Image Registration Techniques
Soumick Chatterjee1,2,3, Himanshi Bajaj3, Suraj Bangalore Shashidhar3, Sanjeeth Busnur Indushekar3, Steve Simon3, Istiyak Hossain Siddiquee3, Nandish Bandi Subbarayappa3, Oliver Speck1,4,5,6, and Andreas Nürnberger2,3,6
1Department of Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4German Centre for Neurodegenerative Diseases, Magdeburg, Germany, 5Leibniz Institute for Neurobiology, Magdeburg, Germany, 6Center for Behavioral Brain Sciences, Magdeburg, Germany
Deep learning algorithms have been used extensively in tackling medical image registration issues. However, these methods have not thoroughly evaluated on datasets representing real clinic scenarios. Hence in this survey, three state-of-the-art methods were compared against the gold standards ANTs and FSL, for performing deformable image registrations on publicly available IXI dataset, which resembles clinical data. The comparisons were performed for intermodality and intramodality registration tasks; though in all the respective papers, only the intermodality registrations were exhibited. The experiments have shown that for intramodality tasks, all the methods performed reasonably well and for intermodality tasks the methods faced difficulties.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.