Digital Posters
Machine Learning: Removing Artifacts & Improving Resolution
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 13:00 - 14:00 |
1764.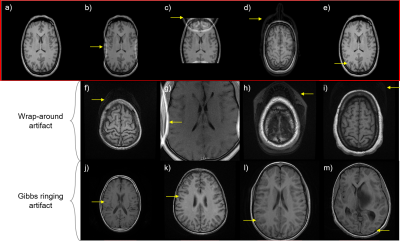 |
ArtifactID: identifying artifacts in low field MRI using deep learning
Marina Manso Jimeno1,2, Keerthi Sravan Ravi2,3, John Thomas Vaughan Jr.2,3, Dotun Oyekunle4, Godwin Ogbole4, and Sairam Geethanath2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States, 3Columbia University, New York, NY, United States, 4Radiology, University College Hospital, Ibadan, Nigeria
Most MR images contain artifacts such as wrap-around and Gibbs ringing, which negatively affect the diagnostic quality and, in some cases, may be confused with pathology. This work presents ArtifactID, a deep learning based tool to help MR technicians to identify and classify artifacts in datasets acquired with low-field systems. We trained binary classification models to accuracies greater than 88% to identify wrap-around and Gibbs ringing artifacts in T1 brain images. ArtifactID can help novice MR technicians in low resource settings to identify and mitigate these artifacts.
|
|||
1765.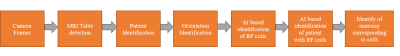 |
Artificial Intelligence based smart MRI: Towards development of automated workflow for reduction of repeat and reject of scans
Raghu Prasad, PhD1, Harikrishna Rai, PhD1, and Sanket Mali1
1GE Healthcare, Bangalore, India
Majority of the pre-scan errors in MRI radiological workflows are due to a) in-appropriate patient positioning, b) incorrect protocol selection for the anatomy to be scanned, c) operator/technologist negligence. In this work, we propose and develop an AI (Artificial Intelligence) based computer vision solution to correct patient positioning errors and reduce the scan time. Our approach relies on identification of RF coil and anatomy of the patient when occluded with coils using a 3D depth camera. Camera based solution has shown significant improvements in some of the critical MRI based workflow such as auto-landmarking, coil/protocol selection and scan range overlay.
|
|||
1766.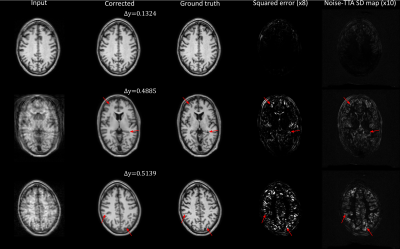 |
Uncertainty estimation for DL-based motion artifact correction in 3D brain MRI
Karsten Sommer1, Jochen Keupp1, Christophe Schuelke1, Oliver Lips1, and Tim Nielsen1
1Philips Research, Hamburg, Germany
Test-time augmentation (TTA) is explored as an uncertainty prediction method for a neural network based 3D motion artifact correction starting from magnitude images. To this end, a synthetic training dataset is generated using a dedicated 3D motion artifact simulation pipeline. After training, a TTA-based uncertainty metric is employed to predict the network performance for data not contained in training. Using synthetic test data, we find that the proposed method can accurately predict the overall motion correction accuracy (total RMSE) but fails in certain cases to reliably detect local “hallucinations” (brain-like structures different from the actual anatomy) of the network.
|
|||
1767.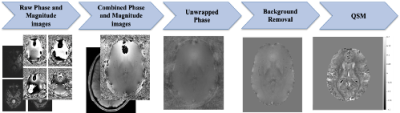 |
A 3D-UNET for Gibbs artifact removal from quantitative susceptibility maps
Iyad Ba Gari1, Shruti P. Gadewar1, Xingyu Wei1, Piyush Maiti1, Joshua Boyd1, and Neda Jahanshad1
1Imaging Genetics Center, Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine of USC, Los Angeles, CA, United States
Magnetic resonance image quality is susceptible to several artifacts including Gibbs-ringing. Although there have been deep learning approaches to address these artifacts on T1-weighted scans, Quantitative susceptibility maps (QSMs), derived from susceptibility-weighted imaging, are often more prone to Gibbs artifacts than T1w images, and require their own model. Removing such artifacts from QSM will improve the ability to non-invasively map iron deposits, calcification, inflammation, and vasculature in the brain. In this work, we develop a 3D U-Net based approach to remove Gibbs-ringing from QSM maps.
|
|||
1768.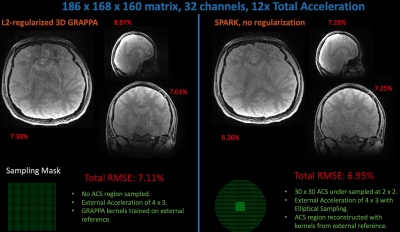 |
Extending Scan-specific Artifact Reduction in K-space (SPARK) to Advanced Encoding and Reconstruction Schemes
Yamin Arefeen1, Onur Beker2, Heng Yu3, Elfar Adalsteinsson4,5,6, and Berkin Bilgic5,7,8
1Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Department of Computer and Communication Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 3Department of Automation, Tsinghua University, Beijing, China, 4Massachusetts Institute of Technology, Cambridge, MA, United States, 5Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 6Institute for Medical Engineering and Science, Cambridge, MA, United States, 7Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 8Department of Radiology, Harvard Medical School, Boston, MA, United States
Scan-specific learning techniques improve accelerated MRI reconstruction by training models using data solely from the specific scan; but they are constrained to Cartesian imaging and require an integrated auto-calibration signal (ACS), reducing acceleration. This abstract extends the scan-specific model SPARK, which estimates and corrects reconstruction errors in k-space, to arbitrary acquisitions and reconstructions. We demonstrate improvements in 3D volumetric imaging either with an integrated or external ACS region and in simultaneous multi-slice, wave-encoded imaging. SPARK enables an order of magnitude acceleration with ~2-fold reduction in reconstruction error compared to advanced reconstruction techniques that serve as its input.
|
|||
1769.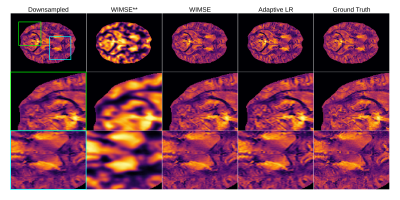 |
Improving Deep Learning MRI Super-Resolution for Quantitative Susceptibility Mapping
Antoine Moevus1,2, Mathieu Dehaes2,3,4, and Benjamin De Leener1,2,4
1Departement of Computer and Software Engineering, Polytechnique Montréal, Montréal, QC, Canada, 2Institute of Biomedical Engineering, University of Montreal, Montréal, QC, Canada, 3Department of Radiology, Radio-oncology, and Nuclear Medicine, Université de Montréal, Montréal, QC, Canada, 4Research Center, Ste-Justine Hospital University Centre, Montreal, QC, Canada
In this preliminary work, we are exploring the application of deep learning (DL) super-resolution techniques to improve quantitative susceptibility maps (QSM). We trained a light deep learning neural network on the QSM data from the AHEAD dataset. We studied different variants of the mean squared error (MSE) as loss functions and two different training strategies : cyclic learning rate and an adaptive learning rate. We found that the cyclic learning rate yielded better results in general if correctly optimized with the learning rate finder algorithm.
|
|||
1770.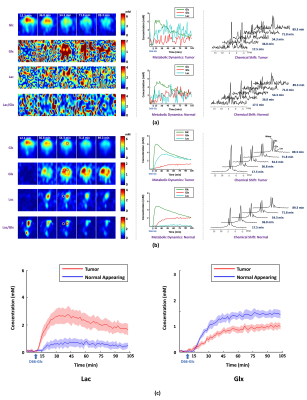 |
A Marriage of Subspace Modeling with Deep Learning to Enable High-Resolution Dynamic Deuterium MR Spectroscopic Imaging
Yudu Li1,2, Yibo Zhao1,2, Rong Guo1,2, Tao Wang3, Yi Zhang3, Mathew Chrostek4, Walter C. Low4, Xiao-Hong Zhu3, Wei Chen3, and Zhi-Pei Liang1,2
1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 4Department of Neurosurgery, University of Minnesota, Minneapolis, MN, United States
Dynamic deuterium MR spectroscopic imaging (DMRSI) is a powerful metabolic imaging method, with great potential for tumor imaging. However, current DMRSI applications are limited to low spatiotemporal resolutions due to low sensitivity. This work overcomes this issue using a machine learning-based method. The proposed method integrates subspace modeling with deep learning to effectively use prior information for sensitivity enhancement and thus enables high-resolution dynamic DMRSI. Experimental results have been obtained from rats with and without brain tumor, which demonstrate that we can obtain dynamic metabolic changes with unprecedented spatiotemporal resolutions.
|
|||
1771.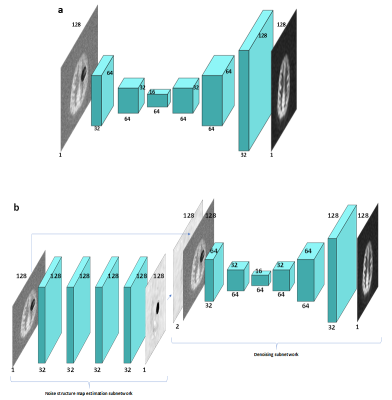 |
Removing structured noise from dynamic arterial spin labeling images
Yanchen Guo1, Zongpai Zhang1, Shichun Chen1, Lijun Yin1, David C. Alsop2, and Weiying Dai1
1Department of Computer Science, State University of New York at Binghamton, Binghamton, NY, United States, 2Department of Radiology, Beth Israel Deaconess Medical Center & Harvard Medical School, Boston, MA, United States
Dynamic arterial spin labeling (dASL) images showed the existence of large-scale structured noise, which violates the Gaussian assumptions of baseline functional imaging studies. Here, we evaluated the performance of two deep neural network (DNN) methods on removing the structured noise of ASL images, using the simulated data and real image data. The DNN model, with the noise structure learned and incorporated, demonstrates consistently improved performance compared to the DNN model without the explicitly incorporated noise structure. These results indicate that the noise structure incorporated DNN model is promising in removing the structured noise from the ASL functional images.
|
|||
1772.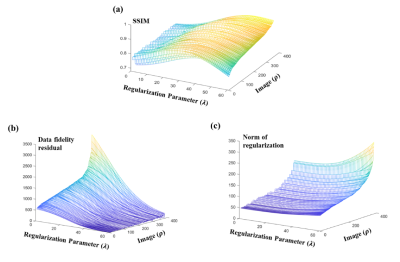 |
Learn to Better Regularize in Constrained Reconstruction
Yue Guan1, Yudu Li2,3, Xi Peng4, Yao Li1, Yiping P. Du1, and Zhi-Pei Liang2
1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Mayo Clinic, Rochester, MN, United States
Selecting good regularization parameters is essential for constrained reconstruction to produce high-quality images. Current constrained reconstruction methods either use empirical values for regularization parameters or apply some computationally expensive test, such as L-curve or cross-validation, to select those parameters. This paper presents a novel learning-based method for determination of optimal regularization parameters. The proposed method can not only determine the regularization parameters efficiently but also yield more optimal values in terms of reconstruction quality. The method has been evaluated using experimental data in three constrained reconstruction scenarios, producing excellent reconstruction results using the selected regularization parameters.
|
|||
 |
1773.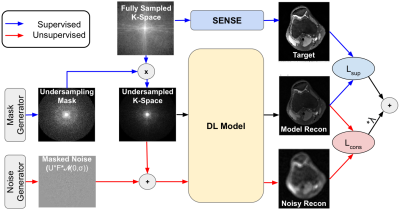 |
Noise2Recon: A Semi-Supervised Framework for Joint MRI Reconstruction and Denoising using Limited Data
Arjun D Desai1,2, Batu M Ozturkler1, Christopher M Sandino3, Brian A Hargreaves2,3,4, John M Pauly3,5, and Akshay S Chaudhari2,5,6
1Electrical Engineering (Equal Contribution), Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Bioengineering, Stanford University, Stanford, CA, United States, 5Equal Contribution, Stanford University, Stanford, CA, United States, 6Biomedical Data Science, Stanford University, Stanford, CA, United States
Deep learning (DL) has shown promise for faster, high quality accelerated MRI reconstruction. However, standard supervised DL methods depend on extensive amounts of fully-sampled, ground-truth data and are sensitive to out-of-distribution (OOD), particularly low-SNR, data. In this work, we propose a semi-supervised, consistency-based framework (termed Noise2Recon) for joint MR reconstruction and denoising that uses a limited number of fully-sampled references. Results demonstrate that even with minimal ground-truth data, Noise2Recon can use unsupervised, undersampled data to 1) achieve high performance on in-distribution (noise-free) scans and 2) improve generalizability to noisy, OOD scans compared to both standard and augmentation-based supervised methods.
|
||
1774.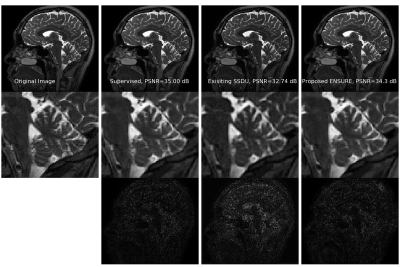 |
ENSURE: Ensemble Stein’s Unbiased Risk Estimator for Unsupervised Learning
Hemant Kumar Aggarwal1, Aniket Pramanik1, and Mathews Jacob1
1Electrical and Computer Engineering, University of Iowa, Iowa City, IA, United States
Deep learning algorithms are emerging as powerful alternatives to compressed sensing methods, offering improved image quality and computational efficiency. Fully sampled training images are often difficult to acquire in high-resolution and dynamic imaging applications. We propose an ENsemble SURE (ENSURE) loss metric to train a deep network only from undersampled measurements. In particular, we show that training a network using an ensemble of images, each acquired with a different sampling pattern, using ENSURE can provide results that closely approach MSE training. Our experimental results show comparable reconstruction quality to supervised learning.
|
|||
1775.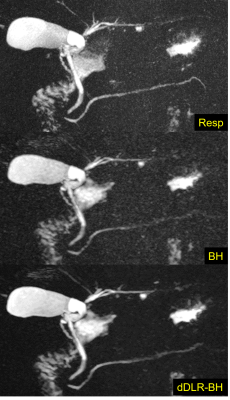 |
Breath-hold 3D MRCP at 1.5T using Fast 3D mode and a deep learning-based noise-reduction technique
Taku Tajima1,2, Hiroyuki Akai3, Koichiro Yasaka3, Rintaro Miyo1, Masaaki Akahane2, Naoki Yoshioka2, and Shigeru Kiryu2
1Department of Radiology, International University of Health and Welfare Mita Hospital, Tokyo, Japan, 2Department of Radiology, International University of Health and Welfare Narita Hospital, Chiba, Japan, 3Department of Radiology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
The usefulness of breath-hold 3D MRCP at 3T MRI has recently been reported. Moreover, the denoising approach with deep learning-based reconstruction (dDLR) is a novel technique. We assessed the image quality of conventional respiratory-triggered 3D MRCP (Resp) and breath-hold 3D MRCP using Fast 3D mode without and with dDLR (BH, dDLR-BH) at 1.5T, by comparing the overall image quality and duct visibility scores. Breath-hold 3D MRCP compared favorably with respiratory-triggered MRCP at 1.5T. dDLR can improve the overall image quality and duct visibility of breath-hold 3D MRCP, and the visibility of the right and left hepatic ducts was improved statistically.
|
|||
1776.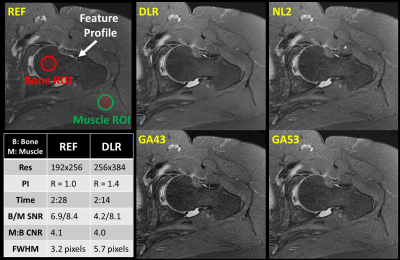 |
Higher Resolution with Improved Image Quality without Increased Scan Time: Is it possible with MRI Deep Learning Reconstruction?
Hung Do1, Carly Lockard2, Dawn Berkeley1, Brian Tymkiw1, Nathan Dulude3, Scott Tashman2, Garry Gold4, Erin Kelly1, and Charles Ho2
1Canon Medical Systems USA, Inc., Tustin, CA, United States, 2Steadman Philippon Research Institute, Vail, CO, United States, 3The Steadman Clinic, Vail, CO, United States, 4Stanford University, Stanford, CA, United States
In magnetic resonance imaging (MRI), increased resolution leads to increased scan time and reduced signal-to-noise ratio (SNR). Parallel imaging (PI) can be used to mitigate the increased scan time but comes with an additional penalty in SNR resulting in reduced image quality. Deep Learning Reconstruction (DLR) has recently been developed to intelligently remove noise from low SNR input images producing increased SNR and quality output images. SNR gain from DLR could be used to increase resolution while maintaining scan time. This work demonstrates that DLR could be used to increase resolution and image quality without increased scan time.
|
|||
1777.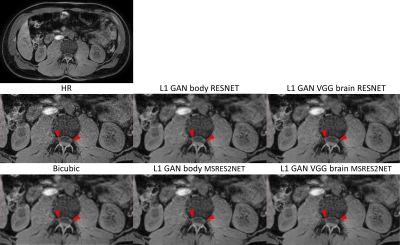 |
Evaluation of Super Resolution Network for Universal Resolution Improvement
Zechen Zhou1, Yi Wang2, Johannes M. Peeters3, Peter Börnert4, Chun Yuan5, and Marcel Breeuwer3
1Philips Research North America, Cambridge, MA, United States, 2MR Clinical Science, Philips Healthcare North America, Gainesville, FL, United States, 3MR Clinical Science, Philips Healthcare, Best, Netherlands, 4Philips Research Hamburg, Hamburg, Germany, 5Vascular Imaging Lab, University of Washington, Seattle, WA, United States
In this work, we investigated whether deep learning based super resolution (SR) network trained from a brain dataset can generalize well to other applications (i.e. knee and abdominal imaging). Our preliminary results imply that 1) the perceptual loss function can improve the generalization performance of SR network across different applications; 2) the multi-scale network architecture can better stabilize the SR results particularly for training dataset with lower quality. In addition, the SR improvement from increased data diversity can be saturated, indicating that a single trained SR network might be feasible for universal MR image resolution improvement.
|
|||
1778.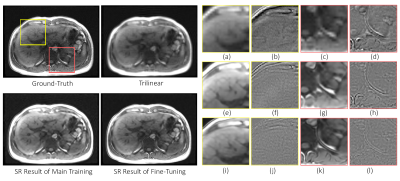 |
Fine-tuning deep learning model parameters for improved super-resolution of dynamic MRI with prior-knowledge
Chompunuch Sarasaen1,2, Soumick Chatterjee1,3,4, Fatima Saad2, Mario Breitkopf1, Andreas Nürnberger4,5,6, and Oliver Speck1,5,6,7
1Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Institute for Medical Engineering, Otto von Guericke University, Magdeburg, Germany, 3Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 4Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
Dynamic imaging is required during interventions to assess the physiological changes. Unfortunately, while achieving a high temporal resolution the spatial resolution is compromised. To overcome the spatiotemporal trade-off, in this work deep learning based super-resolution approach has been utilized and fine-tuned using prior-knowledge. 3D dynamic data for three subjects was acquired with different parameters to test the generalization capabilities of the network. Experiments were performed for different in-plane undersampling levels. A U-net based model[1] with perceptual loss[2] was used for training. Then, the trained network was fine-tuned using prior scan to obtain high resolution dynamic images during the inference stage.
|
|||
1779.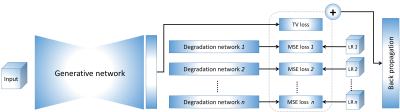 |
MRI super-resolution reconstruction: A patient-specific and dataset-free deep learning approach
Yao Sui1,2, Onur Afacan1,2, Ali Gholipour1,2, and Simon K Warfield1,2
1Harvard Medical School, Boston, MA, United States, 2Boston Children's Hospital, Boston, MA, United States
Spatial resolution is critically important in MRI. Unfortunately, direct high-resolution acquisition is time-consuming and suffers from reduced signal-to-noise ratio. Deep learning-based super-resolution has emerged to improve MRI resolution. However, current methods require large-scale training datasets of high-resolution images, which are difficult to obtain at suitable quality. We developed a deep learning technique that trains the model on the patient-specific low-resolution data, and achieved high-quality MRI at a resolution of 0.125 cubic mm with six minutes of imaging time. Experiments demonstrate our approach achieved superior results to state-of-the-art super-resolution methods, while reduced scan time as delivered with direct high-resolution acquisitions.
|
|||
1780.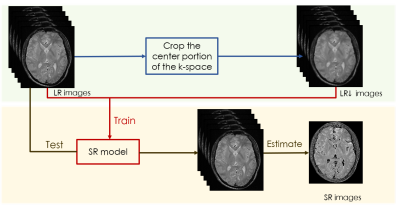 |
An self-supervised deep learning based super-resolution method for quantitative MRI
Muzi Guo1,2, Yuanyuan Liu1, Yuxin Yang1, Dong Liang1, Hairong Zheng1, and Yanjie Zhu1
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Bejing, China
High-resolution (HR) quantitative magnetic resonance images (qMRI) are widely used in clinical diagnosis. However, acquisition of such high signal-to-noise ratio data is time consuming, and could lead to motion artifacts. Super-resolution (SR) approaches provide a better trade-off between acquisition time and spatial resolution. However, State-of-the-art SR methods are mostly supervised, which require external training data consisting of specific LR-HR pairs, and have not considered the quantitative conditions, which leads to the estimated quantitative map inaccurate. An self-supervised super-resolution algorithm under quantitative conditions is presented. Experiments on T1ρ quantitative images show encouraging improvements compared to competing SR methods.
|
|||
1781.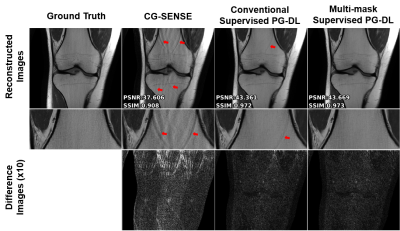 |
Enhancing the Reconstruction quality of Physics-Guided Deep Learning via Holdout Multi-Masking
Burhaneddin Yaman1,2, Seyed Amir Hossein Hosseini1,2, Steen Moeller2, and Mehmet Akçakaya1,2
1University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, Minneapolis, MN, United States
Physics-guided deep learning (PG-DL) approaches unroll conventional iterative algorithms consisting of data consistency (DC) and regularizers, and typically perform training on a fully-sampled database. Although supervised training has been incredibly successful, there is still room for further removing residual and banding artifacts. To improve reconstruction quality and robustness of supervised PG-DL, we propose to use multiple subsets of acquired measurements in the DC units during training by applying a multi-masking operation on available sub-sampled data, unlike existing supervised PG-DL approaches that use all the available measurements in DC units. Proposed method outperforms conventional supervised PG-DL method by further reducing theartifacts.
|
|||
1782.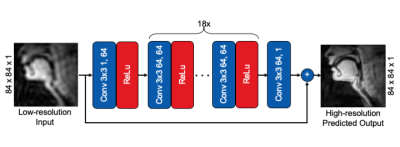 |
Feasibility of Super Resolution Speech RT-MRI using Deep Learning
Prakash Kumar1, Yongwan Lim1, and Krishna Nayak1
1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States Super-resolution using deep learning has been successfully applied to camera imaging and recently to static and dynamic MRI. In this work, we apply super-resolution to the generation of high-resolution real-time MRI from low resolution counterparts in the context of human speech production. Reconstructions were performed using full (ground truth) and truncated zero-padded k-space (low resolution). The network, trained with a common 2D residual architecture, outperformed traditional interpolation based on PSNR, MSE, and SSIM metrics. Qualitatively, the network reconstructed most vocal tract segments including the velum and lips correctly but caused modest blurring of lip boundaries and the epiglottis. |
|||
1783.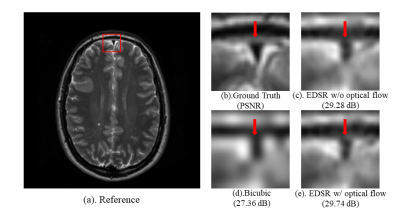 |
Optical Flow-based Data Augmentation and its Application in Deep Learning Super Resolution
Yu He1, Fangfang Tang1, Jin Jin1,2, and Feng Liu1
1School of Information Technology and Electrical Engineering, the University of Queensland, Brisbane, Australia, 2Research and Development MR, Siemens Healthcare, Brisbane, Australia
Deep learning (DL) methods have been a hot topic in MRI reconstruction, such as super-resolution. However, DL usually requires a substantial amount of training data, which may not always be accessible because of limited clinical cases, privacy limitation, the cross-vendor, and cross-scanner variation, etc. In this work, we propose an affine transformation data augmentation method to increase training data for MRI super-resolution. Comprehensive experiments were performed on real T2 brain images to validate the proposed method.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.