Digital Posters
Machine Learning to Reconstruct Accelerated Scans
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 15:00 - 16:00 |
1964.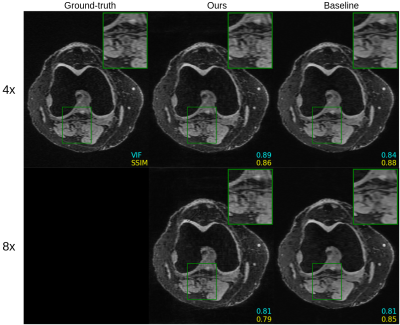 |
Using Untrained Convolutional Neural Networks to Accelerate MRI in 2D and 3D
Dave Van Veen1,2, Arjun Desai1, Reinhard Heckel3,4, and Akshay S. Chaudhari1
1Stanford University, Stanford, CA, United States, 2University of Texas at Austin, Austin, TX, United States, 3Rice University, Houston, TX, United States, 4Technical University of Munich, Munich, Germany
We investigate untrained convolutional neural networks for accelerating both 2D and 3D MRI scans of the knee. Machine learning has demonstrated great potential to accelerate scans while maintaining high quality reconstructions. However, these methods are often trained over a large number of fully-sampled scans, which are difficult to acquire. Here we demonstrate MRI acceleration with untrained networks, achieving similar performance to a trained baseline. Further, we use undersampled k-space measurements as regularization priors to increase the robustness of untrained methods.
|
|||
1965.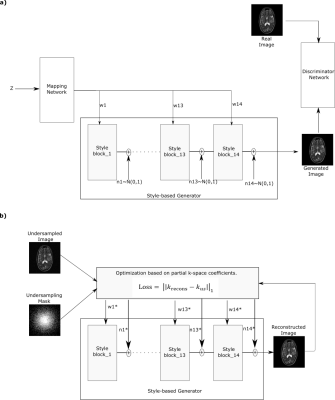 |
Zero-shot Learning for Unsupervised Reconstruction of Accelerated MRI Acquisitions
Yilmaz Korkmaz1,2, Salman Ul Hassan Dar1,2, Mahmut Yurt1,2, and Tolga Çukur1,2,3
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center, Bilkent University, Ankara, Turkey, 3Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey
A popular framework for reconstruction of undersampled MR acquisitions is deep neural networks (DNNs). DNNs are typically trained in a supervised manner to learn mapping between undersampled and fully sampled acquisitions. However, this approach ideally requires training a separate network for each set of contrast, acceleration rate, and sampling density, which introduces practical burden. To address this limitation, we propose a style generative model that learns MR image priors given fully sampled training dataset of specific contrast. Proposed approach is then able to efficiently recover undersampled acquisitions without any training, irrespective of the image contrast, acceleration rate or undersampling pattern.
|
|||
 |
1966.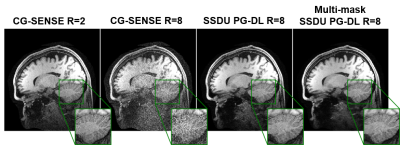 |
Multi-Mask Self-Supervised Deep Learning for Highly Accelerated Physics-Guided MRI Reconstruction
Burhaneddin Yaman1,2, Seyed Amir Hossein Hosseini1,2, Steen Moeller2, Jutta Ellermann2, Kâmil Uğurbil2, and Mehmet Akçakaya1,2
1University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, Minneapolis, MN, United States
Self-supervised physics-guided deep learning (PG-DL) approaches enable training neural networks without fully-sampled data. These methods split the available k-space measurements into two sets. One is used in the data consistency units of the unrolled network, while the other is used to define the loss. Although self-supervised learning performs well at moderately high acceleration rates, scarcity of acquired data at high acceleration rates degrades the reconstruction performance. In this work, we propose a multi-mask self-supervised learning approach, which retrospectively splits acquired measurement into multiple 2-tuples of disjoint sets. Proposed multi-mask self-supervised learning method outperforms its single-mask counterpart at high acceleration rates.
|
||
1967.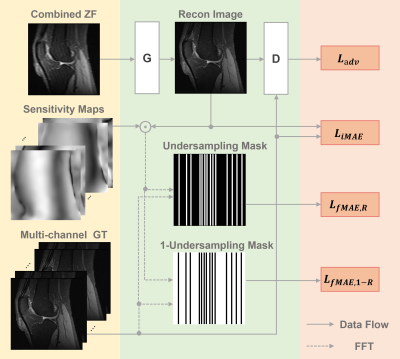 |
PIC-GAN: A Parallel Imaging Coupled Generative Adversarial Network for Accelerated Multi-Channel MRI Reconstruction
Jun Lyu1, Chengyan Wang2, and Guang Yang3,4
1School of Computer and Control Engineering, Yantai University, Yantai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Cardiovascular Research Centre, Royal Brompton Hospital, London, United Kingdom, 4National Heart and Lung Institute, Imperial College London, London, United Kingdom
To demonstrate the feasibility of combining parallel imaging (PI) with the generative adversarial network (GAN) for accelerated multi-channel MRI reconstruction. In our proposed PIC-GAN framework, we used a progressive refinement method in both frequency and image domains, which can not only help to stabilize the optimization of the network, but also make full use of the complementarity of the two domains. More specifically, the loss function in the image domain ensures to reduce aliasing artifacts between the reconstructed images and their corresponding ground truth. This enables the model to ensure high-fidelity reconstructions can be obtained even at high acceleration factors.
|
|||
1968.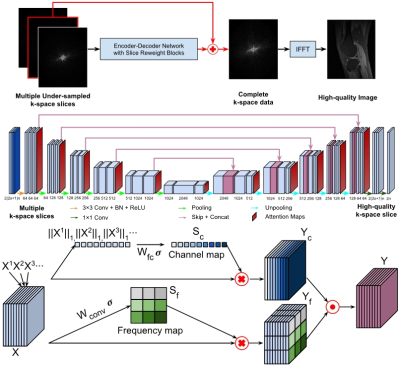 |
Adaptive convolutional neural networks for accelerating magnetic resonance imaging via k-space data interpolation
Tianming Du1, Yuemeng Li1, Honggang Zhang2, Stephen Pickup1, Rong Zhou1, Hee Kwon Song1, and Yong Fan1
1Radiology, University of Pennsylvania, Philadelphia, PA, United States, 2Beijing University of Posts and Telecommunications, Beijing, China
Deep learning in k-space has demonstrated great potential for image reconstruction from undersampled k-space data. However, existing deep learning-based image reconstruction methods typically apply weight-sharing convolutional neural networks to k-space data without taking into consideration the k-space data’s spatial frequency properties, leading to ineffective learning of the image reconstruction models. To improve image reconstruction performance, we develop a residual Encoder-Decoder network architecture with self-attention layers to adaptively focus on k-space data at different spatial frequencies and channels for interpolating the undersampled k-space data. Experimental results demonstrate that our method achieves significantly better image reconstruction performance than current state-of-the-art techniques.
|
|||
1969.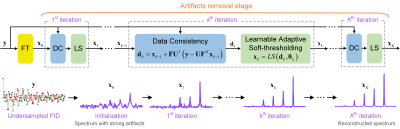 |
Accelerated Magnetic Resonance Spectroscopy with Model-inspired Deep Learning
Zi Wang1, Yihui Huang1, Zhangren Tu2, Di Guo2, Vladislav Orekhov3, and Xiaobo Qu1
1Department of Electronic Science, National Institute for Data Science in Health and Medicine, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 3Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden
Multi-dimensional nuclear magnetic resonance (NMR) spectroscopy is an invaluable biophysical tool but often suffers from long measurement. Several methods have been established for spectra reconstruction from undersampled data, two of which are model-based optimization and data-driven deep learning. Combining the main merits of them, we present a model-inspired flexible deep learning framework, for reliable, robust, and ultra-fast spectra reconstruction. Besides, we demonstrate that the model-inspired network needs very few parameters and is not sensitive to training datasets, which greatly reduces the demand for memory footprints and can work effectively in a wide range of scenarios without re-training.
|
|||
1970.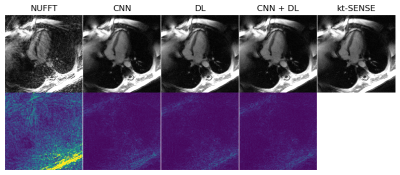 |
DL2 - Deep Learning + Dictionary Learning-based Regularization for Accelerated 2D Dynamic Cardiac MR Image Reconstruction
Andreas Kofler1, Tobias Schaeffter1,2,3, and Christoph Kolbitsch1,2
1Physikalisch-Technische Bundesanstalt, Berlin and Braunschweig, Berlin, Germany, 2School of Imaging Sciences and Biomedical Engineering, King's College London, London, United Kingdom, 3Department of Biomedical Engineering, Technical University of Berlin, Berlin, Germany
In this work, we combine Convolutional Neural Networks (CNN)- with Dictionary Learning (DL)- and Sparse Coding (SC)-based regularization for dynamic cardiac MR image reconstruction. The regularization on the image is imposed by patch-wise sparsity with respect to a learned overcomplete dictionary and closeness to a CNN-based image-prior which is obtained from a pre-trained CNN. We compare the proposed method to two iterative methods which incorporate the different components separately. We demonstrate the combination of CNNs with DL and SC leads to improved image quality and faster convergence compared to DL+SC only.
|
|||
1971.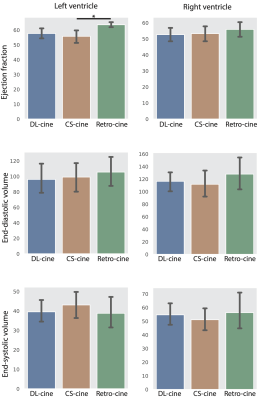 |
Cardiac Functional Analysis with Cine MRI via Deep Learning Reconstruction
Eric Z. Chen1, Xiao Chen1, Jingyuan Lyu2, Qi Liu2, Zhongqi Zhang3, Yu Ding2, Shuheng Zhang3, Terrence Chen1, Jian Xu2, and Shanhui Sun1
1United Imaging Intelligence, Cambridge, MA, United States, 2UIH America, Inc., Houston, TX, United States, 3United Imaging Healthcare, Shanghai, China
Retrospectively gated cine (retro-cine) MRI is the clinical standard for cardiac functional analysis. To the best of our knowledge, this is the first work to evaluate the cine MRI with deep learning reconstruction for cardiac function analysis and compare it with other conventional methods. The cardiac functional values obtained from cine MRI with deep learning reconstruction are consistent with values from clinical standard retro-cine MRI.
|
|||
1972.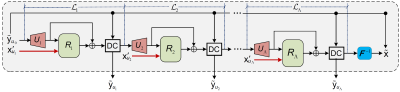 |
Deep Laplacian Pyramid Networks for Fast MRI Reconstruction with Multiscale T1 Priors
Xiaoxin Li1,2, Xinjie Lou1, Junwei Yang3, Yong Chen4, and Dinggang Shen2,5
1College of Computer Science and Technology, Zhejiang University of Technology, Hangzhou, China, 2School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 3Department of Computer Science and Technology, University of Cambridge, Cambridge, United Kingdom, 4Case Western Reserve University, Cleveland, OH, United States, 5Shanghai United Imaging Intelligence Co., Ltd., Shanghai, China
Recent studies explored the merits of using T1-weighted image (T1WI) as a guidance to reconstruct other undersampled MRI modalities via convolutional neural networks (CNNs). However, even aided by T1WI, the reconstruction of highly undersampled MRI data is still suffering from aliasing artifacts, due to the one-upsampling style of existing CNN architectures and not fully using the T1 information. To address these issues, we propose a deep Laplacian pyramid MRI reconstruction framework (LapMRI), which performs progressive upsampling while integrating multiscale prior of T1WI. We show that LapMRI consistently outperforms state-of-the-art methods and can preserve anatomical structure faithfully up to 12-fold undersampling.
|
|||
1973.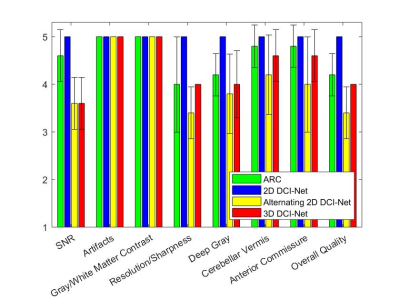 |
Deep learning-based reconstruction of highly-accelerated 3D MRI MPRAGE images
Sangtae Ahn1, Uri Wollner2, Graeme McKinnon3, Rafi Brada2, John Huston4, J. Kevin DeMarco5, Robert Y. Shih5,6, Joshua D. Trzasko4, Dan Rettmann7, Isabelle Heukensfeldt Jansen1, Christopher J. Hardy1, and Thomas K. F. Foo1
1GE Research, Niskayuna, NY, United States, 2GE Research, Herzliya, Israel, 3GE Healthcare, Waukesha, WI, United States, 4Mayo Clinic College of Medicine, Rochester, MN, United States, 5Walter Reed National Military Medical Center, Bethesda, MD, United States, 6Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 7GE Healthcare, Rochester, MN, United States
Three-dimensional (3D) MRI can achieve higher spatial resolution and signal-to-noise ratio than 2D MRI at the expense of long scan times. Recently, deep-learning (DL) techniques have been applied to reconstruction from highly undersampled data, resulting in significant scan accelerations. To assess clinical acceptability, we evaluated DL-based reconstruction on 3D MPRAGE data, using scores from image evaluation by neuroradiologists. Our DCI-Net method with reduction factor R=10 received scores higher than or equal to those of conventional parallel imaging with R=2.1. This implies the DL method can accelerate scans by an additional factor of 5 while maintaining comparable diagnostic image quality.
|
|||
1974.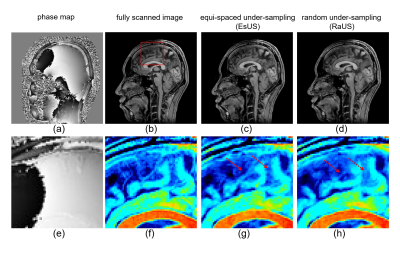 |
Improved CNN-based Image reconstruction using regularly under-sampled signal obtained in phase scrambling Fourier transform imaging
satoshi ITO1 and Shun UEMATSU1
1Utsunomiya University, Utsunomiya, Japan
It has been reported that a quadratic phase scrambling of the spin system in advance to Fourier encoding (PSFT) is effective to the improvement of image quality. In this paper, an CNN-based image reconstruction using PSFT signal was examined. Simulation studies showed that proposed method allows equi-spaced under-sampling and that preservation of structure and image contrast were improved compared to standard Fourier transform based CS-CNN or iterative image reconstruction method. These studies indicate that PSFT has the possibility to reconstruct higher quality images in deep learning image reconstruction as well as iterative reconstruction.
|
|||
1975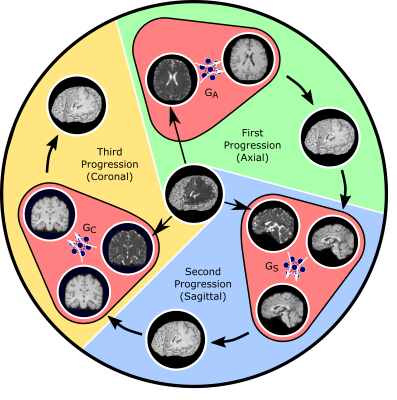 |
Progressive Volumetrization for Data-Efficient Image Recovery in Accelerated Multi-Contrast MRI Video Permission Withheld
Mahmut Yurt1,2, Muzaffer Ozbey1,2, Salman Ul Hassan Dar1,2, Berk Tinaz1,2,3, Kader Karlı Oğuz2,4, and Tolga Çukur1,2,5
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center, Bilkent University, Ankara, Turkey, 3Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 4Department of Radiology, Hacettepe University, Ankara, Turkey, 5Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey
The gold-standard recovery models for accelerated multi-contrast MRI either involve volumetric or cross-sectional processing. Volumetric models offer elevated capture of global context, but may yield suboptimal training due to expanded model complexity. Cross-sectional models demonstrate improved training with reduced complexity, yet may suffer from loss of global consistency in the longitudinal dimension. We propose a novel progressively volumetrized generative model (ProvoGAN) for contextual learning of image recovery in accelerated multi-contrast MRI. ProvoGAN empowers capture of global and local context while maintaining lower model complexity by performing aimed volumetric mappings via a cascade of cross-sectional mappings task-optimally ordered across rectilinear orientations.
|
|||
1976.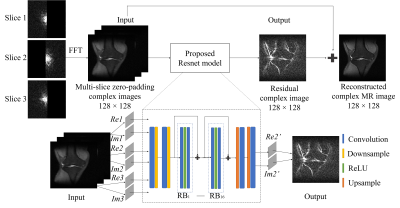 |
Enhanced Multi-Slice Partial Fourier MRI Reconstruction Using Residual Network
Linshan Xie1,2, Yilong Liu1,2, Linfang Xiao1,2, Peibei Cao1,2, Alex T. L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China
In this work, we proposed a residual network based reconstruction method for multi-slice partial Fourier acquisition, where adjacent slices are sampled in a complementary way. The anatomical structure and phase similarity of multi-slice MR data can be exploited to provide complementary information from adjacent slices with different sampling patterns. The proposed method enables highly partial Fourier imaging without noise amplification.
|
|||
1977.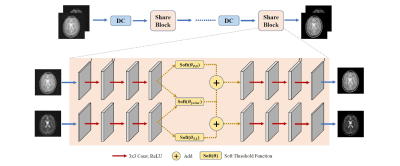 |
Joint-ISTA-Net: A model-based deep learning network for multi-contrast CS-MRI reconstruction
Yuan Lian1, Xinyu Ye1, Yajing Zhang2, and Hua Guo1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 2MR Clinical Science, Philips Healthcare, Suzhou, China Compressed Sensing theory is often applied to accelerate the acquisition of multi-contrast MR images. When highly undersampled, CS-MRI suffers from non-negligible reconstruction error. Here we propose an unrolled iterative deep-learning model to further utilize the group sparsity property for multi-contrast MRI reconstruction at high acceleration factor, named Joint-ISTA-Net, to reduce reconstruction error and aliasing. Our method adds a joint-shrinkage-thresholding model into ISTA-Net to generate a better reconstruction for multi-contrast image pairs. Experiments show the effectiveness of the proposed strategy. |
|||
1978.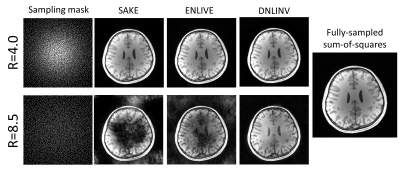 |
Training- and Database-free Deep Non-Linear Inversion (DNLINV) for Highly Accelerated Parallel Imaging and Calibrationless PI&CS MR Imaging
Andrew Palmera Leynes1,2 and Peder E.Z. Larson1,2
1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2UC Berkeley - UC San Francisco Joint Graduate Program in Bioengineering, Berkeley and San Francisco, CA, United States
Deep learning-based MR image reconstruction can provide greater scan time reduction than previously possible. However, this is limited only to MR acquisitions that have large training datasets with fixed hardware and acquisition configurations. We introduce a training- and database-free deep MR image reconstruction technique that may unlock acceleration factors beyond the limits of current state-of-the-art reconstruction methods while being generalizable to any hardware and acquisition configuration. We demonstrate Deep Non-Linear Inversion (DNLINV) on different anatomies and sampling patterns and show high quality reconstructions at higher acceleration factors than previously achievable.
|
|||
1979.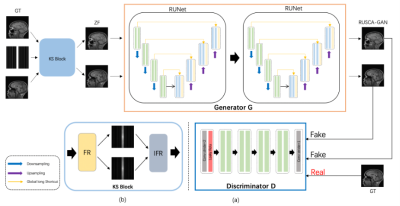 |
A Modified Generative Adversarial Network using Spatial and Channel-wise Attention for Compressed Sensing MRI Reconstruction
Guangyuan Li1, Chengyan Wang2, Weibo Chen3, and Jun Lyu1
1School of Computer and Control Engineering, Yantai University, Yantai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Philips Healthcare, Shanghai, China
At present, many deep learning-based methods have been proposed to solve the problems of traditional CS-MRI, but the reconstruction effect under highly under-sampling has not been well resolved. We proposed a modified GAN architecture for accelerating CS-MRI reconstruction, namely RSCA-GAN. The generator in the proposed architecture is composed of two residual U-net block, in which we added spatial and channel-wise attention (SCA). Each encoding-decoding block is composed of two residual blocks with short skip connections. SCA are added to the decoding block and residual block.
|
|||
1980.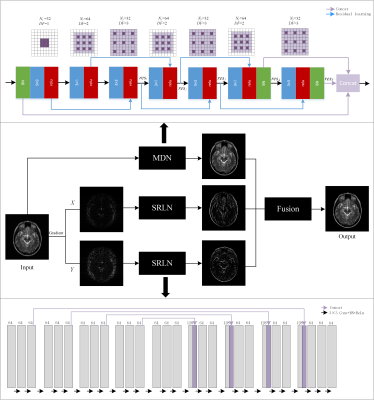 |
Compressed sensing MRI via a fusion model based on image and gradient priors
Yuxiang Dai1, Cheng yan Wang2, and He Wang1
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Human Phenome Institute, Fudan University, Shanghai, China
Compressed sensing (CS) has been employed to accelerate magnetic resonance imaging (MRI) by sampling fewer measurements. We proposed a fusion model based on the optimization method to integrate the image and gradient-based priors into CS-MRI for better reconstruction results via convolutional neural network models. In addition, the proposed fusion model exhibited effective reconstruction performance in magnetic resonance angiography (MRA).
|
|||
1981.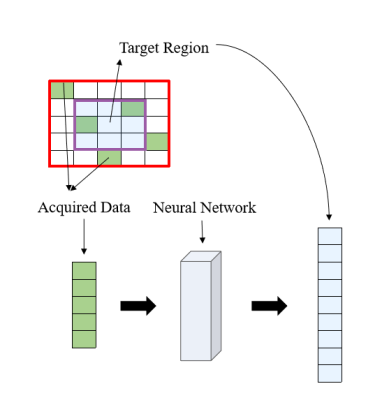 |
Kernel-based Fast EPTI Reconstruction with Neural Network
Muheng Li1, Jie Xiang2, Fuyixue Wang3,4, Zijing Dong3,5, and Kui Ying2
1Department of Automation, Tsinghua University, Beijing, China, 2Department of Engineering Phycics, Tsinghua University, Beijing, China, 3A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 4Harvard-MIT Health Sciences and Technology, MIT, Cambridge, MA, United States, 5Department of Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States
A machine learning based reconstruction framework for Echo Planar Time-resolved Imaging(EPTI) is proposed. This work utilized the special data acquisition trajectory of EPTI, a highly-accelerated spatiotemporal CAIPI sampling, to divide the k-space recovery task into a multi-process program. The missing data is filled within an indicated small kernel with a fully connected neural network. Through image reconstruction tests on human brain data set acquired by EPTI, we demonstrated the high efficiency of this algorithm by shortening the reconstruction time of 216×216×48×32 k-data from over 10 minutes to about 20 seconds.
|
|||
1982.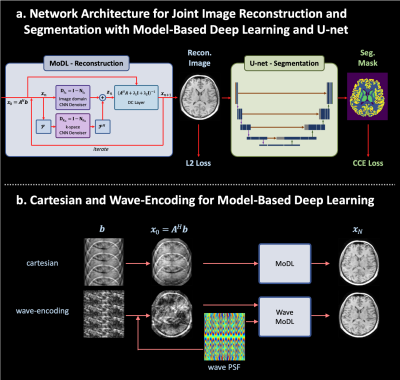 |
Wave-Encoded Model-Based Deep Learning with Joint Reconstruction and Segmentation
Jaejin Cho1,2, Qiyuan Tian1,2, Robert Frost1,2, Itthi Chatnuntawech3, and Berkin Bilgic1,2,4
1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Havard Medical School, Cambridge, MA, United States, 3National Nanotechnology Center, Pathum Thani, Thailand, 4Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
We propose a wave-encoded model-based deep learning (wave-MoDL) strategy for simultaneous image reconstruction and segmentation. We use CNN-based regularizers in both k- and image-space to employ features in both domains and successfully incorporated wave-encoding strategy into MoDL reconstruction. Wave-MoDL enables RyxRz=4x4-fold accelerated 3D imaging using a 32-channel array while reducing NRMSE by 1.45-fold compared to wave-CAIPI. Further, we jointly train wave-MoDL and a U-net for simultaneous reconstruction and segmentation to get additional gain.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.