Digital Posters
Machine Learning Applications in CV Imaging II
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 2 | 13:00 - 14:00 |
 |
2653.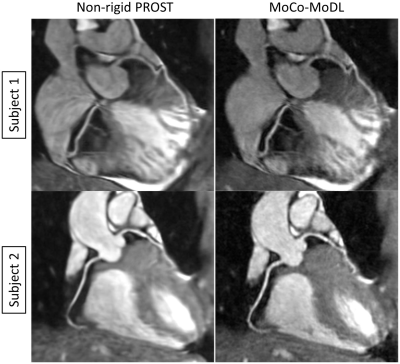 |
End-to-end Motion Corrected Reconstruction using Deep Learning for Accelerated Free-breathing Cardiac MRI
Haikun Qi1, Gastao Cruz1, Thomas Kuestner1, Karl Kunze2, Radhouene Neji2, René Botnar1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
A non-rigid respiratory motion-corrected reconstruction technique (non-rigid PROST) has achieved high-quality coronary MRA (CMRA). However, non-rigid PROST requires respiratory-resolved (bin) image reconstruction, bin-to-bin non-rigid registration and regularized reconstruction, leading to long computation time. In this study, we propose an end-to-end deep learning non-rigid motion-corrected reconstruction technique for highly undersampled free-breathing CMRA. It consists of a diffeomorphic motion estimation network and a motion-informed model-based deep learning reconstruction network that were trained jointly for motion-corrected undersampled reconstruction. Compared with non-rigid PROST, the proposed technique achieved better reconstruction performance in both retrospectively and prospectively 9x-accelerated CMRA, while operating orders of magnitude faster.
|
||
 |
2654.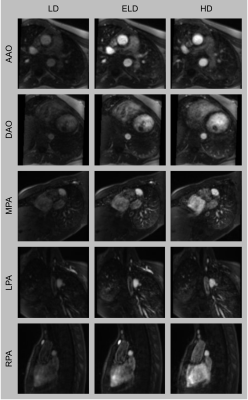 |
Reduction of contrast agent dose in cardiovascular MR angiography using deep learning
Javier Montalt-Tordera1, Michael Quail1, Jennifer Anne Steeden1, and Vivek Muthurangu1
1Centre for Cardiovascular Imaging, UCL Institute of Cardiovascular Science, University College London, London, United Kingdom
Contrast-enhanced MR angiography (CE-MRA) is often used in cardiovascular MRI, but contrast agents can have adverse effects. This work proposes to use deep learning to reduce contrast dose by 80%. A deep neural network was trained to enhance low-dose images using a synthetically generated dataset and validated with both synthetic and real low-dose images. The method was assessed for image quality, diagnostic accuracy and confidence and accuracy of vessel measurements. Enhanced low-dose images were found to be comparable to reference high-dose data. Therefore, the method could enable a reduction in contrast dose for CMR.
|
||
 |
2655.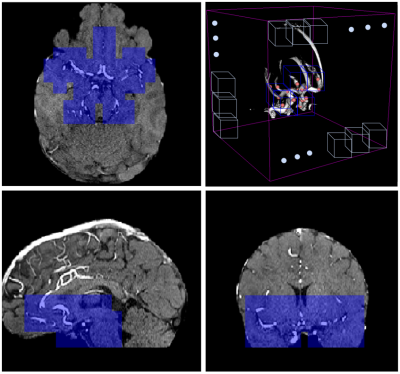 |
Improving automated aneurysm detection on multi-site MRA data: lessons learnt from a public machine learning challenge
Tommaso Di Noto1, Guillaume Marie1, Sebastien Tourbier1, Yasser Alemán-Gómez1,2, Oscar Esteban1, Guillaume Saliou1, Meritxell Bach Cuadra1,3, Patric Hagmann1, and Jonas Richiardi1
1Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 2Center for Psychiatric Neuroscience, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Medical Image Analysis Laboratory (MIAL), Centre d’Imagerie BioMédicale (CIBM), Lausanne, Switzerland
Machine learning challenges serve as a benchmark for determining state-of-the-art results in medical imaging. They provide direct comparisons between algorithms, and realistic estimates of generalization capability. By participating in the Aneurysm Detection And segMentation (ADAM) challenge, we learnt the most effective deep learning design choices to adopt when tackling automated brain aneurysm detection on multi-site data. Adjusting patch overlap ratio during inference, using a hybrid loss, resampling to uniform voxel spacing, using a 3D neural network architecture, and correcting for bias field were the most effective. We show that, when adopting these expedients, our model drastically improves detection performances.
|
||
2656.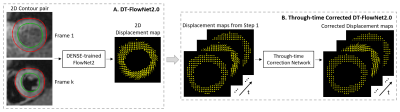 |
Cardiac MRI feature tracking by deep learning from DENSE data
Yu Wang1, Sona Ghadimi1, Changyu Sun1, and Frederick H. Epstein1,2
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Radiology, University of Virginia, Charlottesville, VA, United States
As cine DENSE provides myocardial contours and intramyocardial displacement data, we investigated the use of DENSE to train deep networks to predict intramyocardial motion from contour motion. FlowNet2, an optical-flow convolutional neural network, was used as a comparator/reference, and as the starting point for a DENSE-trained network (DT-FlowNet2). Further, we added a correction network with convolution along time, resulting in a through-time-corrected DENSE-trained network (TC-DT-FlowNet2). TC-DT-FlowNet2 outperformed other methods, providing accurate intramyocardial displacements from myocardial contours. DENSE-based learning of intramyocardial displacements from contours holds promise as a new method for computing strain from the contours of standard cine MRI.
|
|||
2657.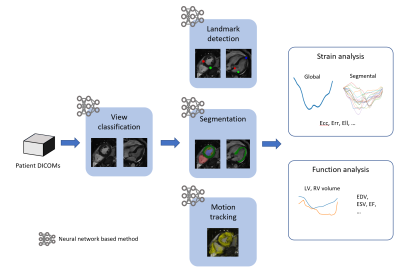 |
Fully Automated Myocardium Strain Analysis using Deep Learning
Xiao Chen1, Masoud Edalati2, Qi Liu2, Xingxian Shou2, Abhishek Sharma1, Mary P. Watkins3, Daniel J. Lenihan3, Linzhi Hu2, Gregory M. Lanza3, Terrence Chen1, and Shanhui Sun1
1United Imaging Intelligence, Cambridge, MA, United States, 2UIH America, Inc., Houston, TX, United States, 3Cardiology, Washington University School of Medicine, St. Louis, MO, United States
Myocardium strain measures myocardial deformation and has been demonstrated a comprehensive, sensitive and early indicator of cardiac dysfunction. Feature tracking can assess myocardium strain from cine CMR images with no special acquisitions but requires observer intervention and expertise. We propose a deep-learning-based fully-automated myocardium strain assessment system that requires zero human intervention for accurate strain assessment.
|
|||
2658.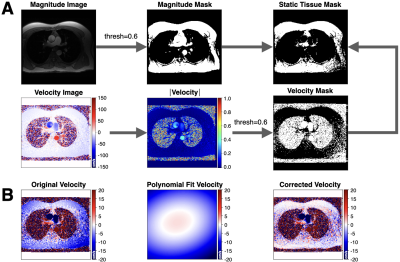 |
Deep Learning-Based ESPIRiT Reconstruction for Accelerated 2D Phase Contrast MRI: Analysis of the Impact of Reconstruction Induced Phase Errors
Matthew J. Middione1, Julio A. Oscanoa1,2, Michael Loecher1, Christopher M. Sandino3, Shreyas S. Vasanawala1, and Daniel B. Ennis1,4
1Department of Radiology, Stanford University, Palo Alto, CA, United States, 2Department of Bioengineering, Stanford University, Palo Alto, CA, United States, 3Department of Electrical Engineering, Stanford University, Palo Alto, CA, United States, 4Cardiovascular Institute, Stanford University, Stanford, CA, United States
2D PC-MRI Compressed sensing (CS) and deep learning (DL) reconstruction techniques may introduce a reconstruction induced phase bias, distinct from eddy current-induced background phase offsets, which may impact the accuracy of flow measurements if not corrected. Herein, we analyzed this reconstruction induced phase bias to determine the maximum acceleration factor that could be used with CS and DL reconstruction frameworks for 2D PC-MRI while minimizing errors in peak velocity and total flow within ±5%.
|
|||
2659.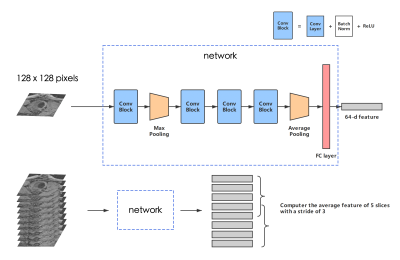 |
Exploring feature space of MR vessel images with limited data annotations through metric learning and episodic training
Kaiyue Tao1, Li Chen2, Niranjan Balu3, Gador Canton3, Wenjin Liu3, Thomas S. Hatsukami4, and Chun Yuan3
1University of Science and Technology of China, Hefei, China, 2Department of Electrical and Computer Engineering, University of Washington, Seattle, WA, United States, 3Department of Radiology, University of Washington, Seattle, WA, United States, 4Department of Surgery, University of Washington, Seattle, WA, United States
Popliteal vessel wall features hidden in the Osteoarthritis Initiative (OAI) dataset warrant further investigation. However, if the number of annotations is insufficient, deep learning-based feature map analysis from MRI images may overfit and fail to generate a meaningful feature space. We designed a metric learning network combined with an episodic training method to overcome the problem of limited annotations, and demonstrated its ability to learn a meaningful feature embedding. Based on our feature map, we proposed an iterative workflow and identified vessel wall images with high probability of diseases from 1974 cases.
|
|||
2660.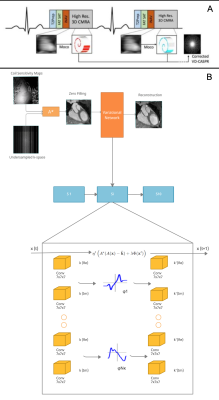 |
Deep learning-based reconstruction for 3D coronary MR angiography with a 3D variational neural network (3D-VNN)
Ioannis Valasakis1, Haikun Qi1, Kerstin Hammernik2, Gastao Lima da Cruz1, Daniel Rueckert2,3, Claudia Prieto1, and Rene Botnar1
1King's College London, London, United Kingdom, 2Technical University of Munich, Munich, Germany, 3Imperial College London, London, United Kingdom
3D whole-heart coronary MR angiography (CMRA) is limited by long scan times. Undersampled reconstruction approaches, such as compressed sensing or low-rank methods show promise to significantly accelerate CMRA but are computationally expensive, require careful parameter optimisation and can suffer from residual aliasing artefacts. A 2D multi-scale variational network (VNN) as recently proposed to improve image quality and significantly shorten the reconstruction time. We propose to extend the VNN reconstruction to 3D to fully capture the spatial redundancies in 3D CMRA. The 3D-VNN is compared against conventional and 3D model-based U-Net reconstruction techniques, showing promising results while shortening the reconstruction time.
|
|||
2661.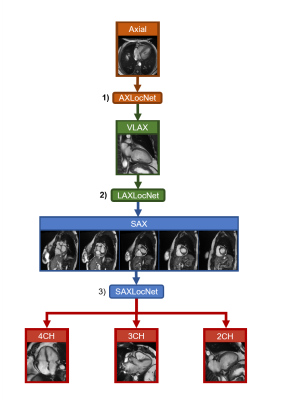 |
Generalizability and Robustness of an Automated Deep Learning System for Cardiac MRI Plane Prescription
Kevin Blansit1, Tara Retson1, Naeim Bahrami2, Phillip Young3,4, Christopher Francois3, Lewis Hahn1, Michael Horowitz1, Seth Kligerman1, and Albert Hsiao1
1UC San Diego, La Jolla, CA, United States, 2GE Healthcare, Menlo Park, CA, United States, 3Mayo Clinic, Rochester, MN, United States, 4Mayo, Rochester, MN, United States
We show that an automated system of deep convolutional neural networks can effectively prescribe double-oblique imaging planes necessary for acquisition of cardiac MRI. To examine its clinical potential, we assess its component-wise performance by comparing simulated imaging planes predicted by DCNNs against ground truth imaging planes defined by a cardiac radiologist. Performance was assessed on 280 exams including 1252 image series obtained on ten 1.5T and 3T MRI scanners from three academic institutions. We further compare imaging planes acquired by technologists at the time of original acquisition against the same ground truth as an additional external reference.
|
|||
2662.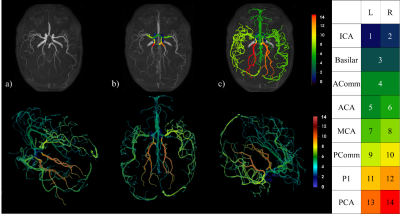 |
Automatic multilabel segmentation of large cerebral vessels from MR angiography images using deep learning
Félix Dumais1, Marco Perez Caceres1, Noémie Arès-Bruneau2, Christian Bocti2,3,4, and Kevin Whittingstall5
1Médecine nucléaire et radiobiologie, Université de Sherbrooke, Sherbrooke, QC, Canada, 2Faculté de Médecine et des Sciences de la Santé, Université de Sherbrooke, Sherbrooke, QC, Canada, 3Clinique de la Mémoire et Centre de Recherche sur le Vieillissement, CIUSSS de l’Estrie-CHUS, Sherbrooke, QC, Canada, 4Service de Neurologie, Département de Médecine, CHUS, Sherbrooke, QC, Canada, 5Radiologie diagnostique, Université de Sherbrooke, Sherbrooke, QC, Canada
The Circle of Willis (CW) is a system of arteries located at the base of the brain. Its structure is a key component in different cerebrovascular diseases, though quantifying this on MR images is meticulous and labor intensive. We developed an analysis pipeline that uses a convolutional neural network to do multilabel segmentation of the CW as viewed through TOF-MRA images. Results show that this approach can automatically locate and label the CW with the same accuracy as expert human annotators.
|
|||
2663.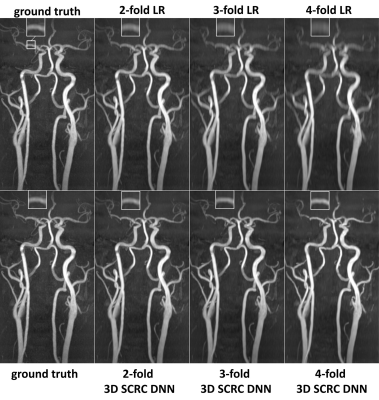 |
Probing the Feasibility and Performance of Super-Resolution Head and Neck MRA Using Deep Machine Learning
Ioannis Koktzoglou1,2, Rong Huang1, William J Ankenbrandt1,2, Matthew T Walker1,2, and Robert R Edelman1,3
1Radiology, NorthShore University HealthSystem, Evanston, IL, United States, 2University of Chicago Pritzker School of Medicine, Chicago, IL, United States, 3Northwestern University Feinberg School of Medicine, Chicago, IL, United States
Deep machine learning approaches offer the potential for improved super-resolution (SR) reconstruction which could be useful in many clinical applications. Patients with suspected stroke often undergo MRI, which often includes magnetic resonance angiography (MRA) of the head and neck arteries with scan times of ≈10 to 15 minutes using standard nonenhanced methods. With the aim of shortening scan times, we evaluated the feasibility and performance of four deep neural network (DNN)-based SR reconstructions for restoring the image quality and spatial resolution of thin slab stack-of-stars quiescent interval slice-selective (QISS) head and neck MRA with degraded slice resolution.
|
|||
2664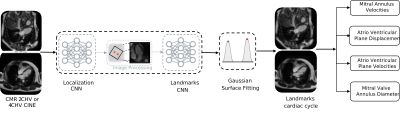 |
Fully automatic extraction of mitral valve annulus motion parameters on long axis CINE CMR using deep learning Video Permission Withheld
Maria Monzon1,2, Seung Su Yoon1,2, Carola Fischer2, Andreas Maier1, Jens Wetzl2, and Daniel Giese2
1Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany
The analysis of mitral valve motion is known to be relevant in the diagnosis of cardiac dysfunction. Dynamic motion parameters can be extracted from Cardiac Magnetic Resonance (CMR) images. We propose two chained Convolutional Neural Networks for automatic tracking of mitral valve-annulus landmarks on time-resolved 2-chamber and 4-chamber CMR images. The first network is trained to detect the region of interest and the second to track the landmarks along the cardiac cycle. We successfully extracted several motion-related parameters with high accuracy as well as analyzed unlabeled datasets, thereby overcoming time-consuming annotation and allowing statistical analysis over large number of datasets.
|
|||
2665.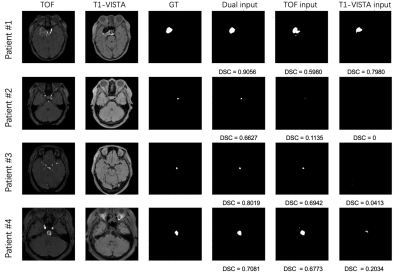 |
Intracranial aneurysm segmentation using a deep convolutional neural network
Miaoqi Zhang1, Qingchu Jin2, Mingzhu Fu1, Hanyu Wei1, and Rui Li1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Johns Hopkins University, Baltimore, MD, United States
Intracranial aneurysms are abnormal dilations of the cerebral arteries that have a prevalence of 5-8% in the general population. In this study, we successfully segmented IAs from dual inputs (TOF-MRA and T1-VISTA) using the hyperdense net with higher accuracy than a single input. The maximum diameter measurements for IAs derived from our segmentation was consistent with the maximum diameters obtained from the criterion standard (DSA). We showed that larger aneurysms were easier to segment by the deep learning model. In the future, we will test other deep learning models on aneurysm segmentation and compare these results with the hyperdense net.
|
|||
2666.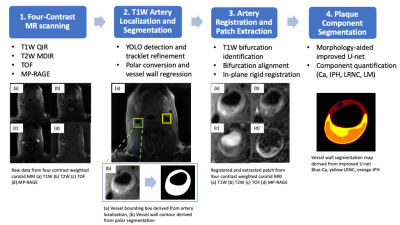 |
AI-based Computer-Aided System for Cardiovascular Disease Evaluation (AI-CASCADE) for carotid tissue quantification
Yin Guo1, Li Chen2, Dongxiang Xu3, Rui Li4, Xihai Zhao4, Thomas S. Hatsukami5, and Chun Yuan1,3
1Bioengineering, University of Washington, Seattle, WA, United States, 2Electrical Engineering, University of Washington, Seattle, WA, United States, 3Radiology, University of Washington, Seattle, WA, United States, 4Biomedical Engineering, Tsinghua University, Beijing, China, 5Surgery, University of Washington, Seattle, WA, United States
The tissue composition of carotid atherosclerotic plaques is crucial for cardiovascular risk assessment and can be quantified with high-resolution multi-contrast MRI by expert reviewers. The purpose of this work is to develop AI-CASCADE, a fully automated solution for quantitative analysis of carotid MRI, including artery localization, vessel wall segmentation, artery registration and plaque component segmentation. Results in our preliminary study show that AI-CASCADE achieves good agreements with manual results and has great potential as an efficient and reliable clinical tool.
|
|||
2667.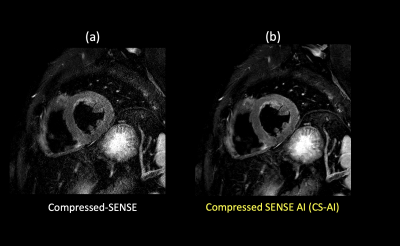 |
Myocardial T2-weighted black-blood imaging with a deep learning constrained Compressed SENSE reconstruction
KOHEI YUDA1, Takashige Yoshida1, Yuki Furukawa1, Masami Yoneyama2, Jihun Kwon2, Nobuo Kawauchi1, Johannes M. Peeters 3, and Marc Van Cauteren3
1Radiology, Tokyo Metropolitan Police Hospital, nakanoku, Japan, 2Philips Japan, Tokyo, Japan, shinagwaku, Japan, 3Philips Healthcare, Best, Netherlands, Netherlands, Netherlands
In high-resolution dual inversion recovery myocardial T2-weighted black-blood (T2W-BB) imaging, using very high acceleration factors with very high resolution can result in degradation of image quality due to insufficient noise removal. In this study, we applied the Compressed-SENSE Artificial Intelligence (CS-AI) framework to further increase the spatial resolution and reduce the scan time. The purpose of this study was to acquire high-resolution myocardial T2W-BB with reduced scan time and compare the image quality between images reconstructed with CS-AI and conventional C-SENSE.
|
|||
2668.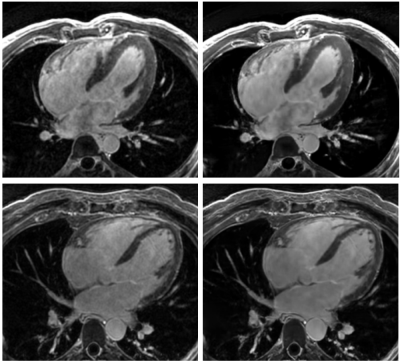 |
Evaluation of a Deep Learning reconstruction framework for three-dimensional cardiac imaging
Gaspar Delso1, Marc Lebel2, Suryanarayanan Kaushik2, Graeme McKinnon2, Paz Garre3, Pere Pujol3, Daniel Lorenzatti3, José T Ortiz3, Susanna Prat3, Adelina Doltra3, Rosario J Perea3, Teresa M Caralt3, Lluis Mont3, and Marta
Sitges3
1GE Healthcare, Barcelona, Spain, 2GE Healthcare, Waukesha, WI, United States, 3Hospital Clínic de Barcelona, Barcelona, Spain
In this study we evaluate a Deep Learning reconstruction framework with adjustable noise reduction on a database of clinical cases, including Delayed Myocardial Enhancement (MDE), Phase-Sensitive MDE (PSIR) and 3D Heart datasets.
|
|||
2669.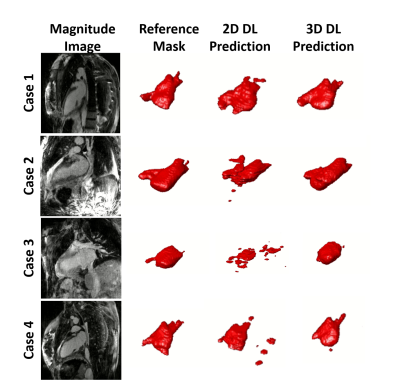 |
Automated Segmentation of the Left Atrium from 3D Late Gadolinium Enhancement Imaging using Deep Learning
Suvai Gunasekaran1, Julia Hwang1, Daming Shen1,2, Aggelos Katsaggelos1,3, Mohammed S.M. Elbaz1, Rod Passman4, and Daniel Kim1,2
1Radiology, Northwestern University, Feinberg School of Medicine, Chicago, IL, United States, 2Biomedical Engineering, Northwestern University, Evanston, IL, United States, 3Electrical and Computer Engineering, Northwestern University, Evanston, IL, United States, 4Cardiology, Northwestern University, Feinberg School of Medicine, Chicago, IL, United States
Left atrial (LA) late gadolinium enhancement (LGE) imaging is essential for detecting fibrosis in patients with atrial fibrillation. Unfortunately, slow manual segmentation of LA LGE limits its use in the clinic. The purpose of this study was to develop a fully automated segmentation method for LA LGE images with deep learning. We tested two different U-net architectures that used either 2D or 3D image inputs for training. Our results demonstrate that 3D inputs are superior to 2D, and the 3D U-Net is a promising method to explore further for clinical translation of LA LGE fibrosis quantification.
|
|||
2670.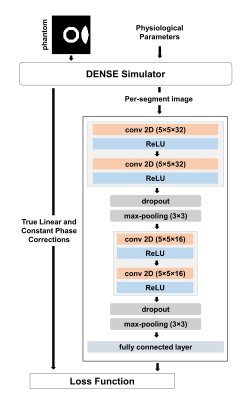 |
Respiratory motion in DENSE MRI: Introduction of a new motion model and use of deep learning for motion correction
Mohamad Abdi1, Daniel S Weller1,2, and Frederick H Epstein1,3
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Electrical and Computer Engineering, University of Virginia, Charlottesville, VA, United States, 3Radiology, University of Virginia, Charlottesville, VA, United States
Conventionally in MRI, respiratory motion leads to shifts of tissue position in the image domain that correspond to linear phase errors in the k-space domain. For DENSE, in addition to position shifts, respiratory motion is displacement-encoded in the stimulated echo, leading to a constant phase error in the k-space domain. We show that in segmented DENSE acquisitions, motion compensation can be applied using per-segment linear and constant phase corrections. As constant phase corrections using image-based navigators are challenging, we show that deep leaning is potentially an effective solution using simulated training data.
|
|||
2671.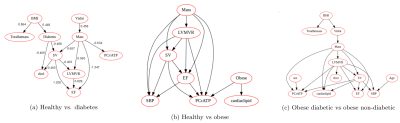 |
Cardiac metabolism assessed by MR Spectroscopy to classify the diabetic and obese heart: a Random Forest and Bayesian network study
Ina Hanninger1, Eylem Levelt2,3, Jennifer J Rayner2, Christopher T Rodgers2,4, Stefan Neubauer2, Vicente Grau1, Oliver J Rider2, and Ladislav Valkovic2,5
1Oxford Institute of Biomedical Engineering, Oxford, United Kingdom, 2Radcliffe Department of Medicine, University of Oxford Centre for Clinical Magnetic Resonance Research, Oxford, United Kingdom, 3University of Leeds, Leeds, United Kingdom, 4Wolfson Brain Imaging Centre, Cambridge, United Kingdom, 5Slovak Academy of Sciences, Institute of Measurement Science, Bratislava, Slovakia
In this study, Random Forest classification was used on data from 197 subjects to discriminate between non-diabetic, diabetic, and obese patients using 31P-MRS and 1H-MRS measurements of cardiac energetics, along with MRI measures of cardiac function. Achieving 91.67%, 73.08% and 88.89% test accuracies, SHAP feature importances indicate a higher predictive impact of metabolic metrics for classifying the diabetic heart compared to global function metrics. Bayesian networks generated through structure learning of the data further suggests a potential causal association of increased visceral fat, increased LVMass resulting in decreased PCr/ATP, and increased cardiac lipid levels attributed to these disease states.
|
|||
2672.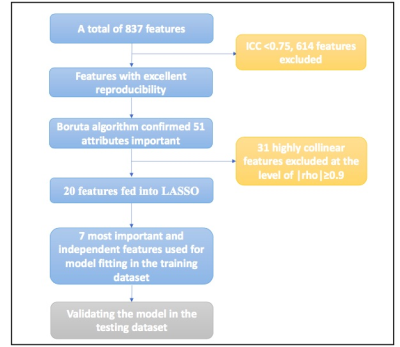 |
Differentiation between cardiac amyloidosis and hypertrophic cardiomyopathy by texture analysis of T2-weighted CMR imaging
Shan Huang1, Yuan Li1, Ke Shi1, Yi Zhang1, Ying-kun Guo2, and Zhi-gang Yang1
1Radiology, West China Hospital, Chengdu, China, 2Radiology, West China Second University Hospital, Chengdu, China
We retrospectively included 100 cardiac amyloidosis (CA) and 217 hypertrophic cardiomyopathy (HCM) patients, aiming to elucidate the value of texture analysis (TA) in non-contrast T2-weighted CMR images of these patients. After the texture features were extracted, machine learning algorithms were used to select the optimal features. The results showed that TA was feasible and reproducible for detecting myocardial tissue alterations and differentiating CA from HCM, even in patients with similar hypertrophy. The radiomics model achieved a comparable diagnostic capacity to late gadolinium enhancement (LGE). Thus, TA might help eliminate the use of contrast agent in the diagnosis of these patients.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.