Digital Posters
Artificial Intelligence
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 4 | 17:00 - 18:00 |
 |
4045.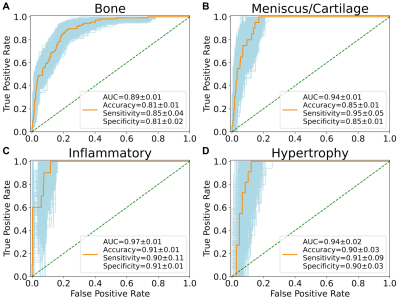 |
Characterizing Knee Osteoarthritis Progression with Structural Phenotypes using MRI and Deep Learning
Nikan K Namiri1, Jinhee Lee1, Bruno Astuto1, Felix Liu1, Rutwik Shah1, Sharmila Majumdar1, and Valentina Pedoia1
1Department of Radiology and Biomedical Imaging and Center for Intelligent Imaging, University of California, San Francisco, San Francisco, CA, United States
We built an end-to-end deep learning model to rapidly stratify knees into morphological phenotypes using a large, longitudinal cohort with knee osteoarthritis (OA). We examined associations of phenotypes with odds of concurrent OA and OA progression. Bone, meniscus/cartilage, and inflammatory phenotypes were strongly associated with current structural OA and symptomatic OA. Hypertrophy phenotype was only weakly associated with structural OA. Among those who did not have baseline OA, bone and meniscus/cartilage phenotypes were strongly associated with developing both structural and symptomatic OA in 48 months. Only bone phenotype increased risk of undergoing total knee replacement surgery within 96 months.
|
||
4046.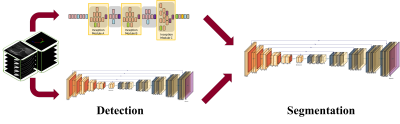 |
Development of Deep Learning based Cartilage Segmentation at 3D knee MRI for the use of Biomarker of Osteoarthritis
Jinwoo Han1, Suk-Joo Hong1, Zepa Yang1, Woo Young Kang1, Yoonmi Choi1, Chang Ho Kang2, Kyung-sik Ahn2, Baek Hyun Kim3, and Euddeum Shim3
1Radiology, Korea University Guro Hospital, KUGH-MIDC, Seoul, Korea, Republic of, 2Korea University Anam Hospital, Seoul, Korea, Republic of, 3Korea University Ansan Hospital, Ansan, Korea, Republic of Cartilage loss is fundamental pathology of knee osteoarthritis (OA). Quantitative analysis of cartilage thickness and volume is very time consuming by manual measurement. We proposed development of deep learning based cartilage segmentation at three dimensional knee magnetic resonance images, which can measure thickness and volume of knee joint cartilage, automatically and accurately. To evaluate the performance, we used Dice Similarity Coefficient (DSC) respect to the manual segmentation, and visual inspection. The accuracy DSC values were higher than 0.9. We expect deep learning program can be useful in future study for knee joint osteoarthritis. |
|||
 |
4047.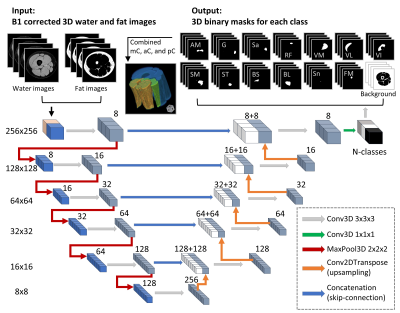 |
Automation of Quantifying Axonal Loss in Patients with Peripheral Neuropathies through Deep Learning Derived Muscle Fat Fraction
Yongsheng Chen1, Daniel Moiseev1, Wan Yee Kong1, Alexandar Bezanovski1, and Jun Li1,2
1Department of Neurology, Wayne State University School of Medicine, Detroit, MI, United States, 2John D. Dingell VA Medical Center, Detroit, MI, United States
Axonal loss determines the final disability in patients with peripheral neuropathies. Consequently, axonal loss results in intramuscular fat accumulation. Therefore, measuring muscle fat fraction through Dixon MRI has been a promising biomarker for monitoring disease progression. However, the responsiveness is yet to be improved, particularly in the early phase of the disease. In this study, we developed a deep learning-based method to automate the quantification of individual muscle fat fraction, which mitigates the laborious manual segmentations and enables the use of individual muscle fat fraction as outcome measures to track axonal loss in patients with neuropathies.
|
||
4048.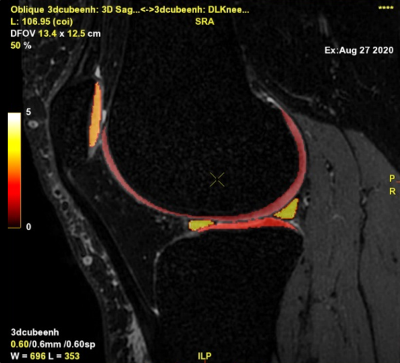 |
Assessment of the potential of a Deep Learning Knee Segmentation and Anomaly Detection Tool in the clinical routine
Laura Carretero1, Pablo García-Polo1, Suryanarayanan Kaushik 2, Maggie Fung2, Bruno Astuto3,4, Rutwik Shah3,4, Pablo F Damasceno3,4, Valentina Pedoia3,4, Sharmila Majumdar3,4, and Mario Padrón5
1Global Research Organization, GE Healthcare, Madrid, Spain, 2GE Healthcare, Waukesha, WI, United States, 3Department of Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 4Center for Digital Health Innovation, UCSF, San Francisco, CA, United States, 5Department of Radiology, Clínica Cemtro, Madrid, Spain
This study evaluates the clinical accuracy of a deep learning (DL)-based tool to segment articular cartilage and menisci on 50 knee MRI exams; detect lesions and stage its severity. An experienced MSK radiologist assessed independently the images for the presence of any lesions on the different compartments and checked the accuracy of its segmentation, resulting in no disagreement with the segmentation output in 92.8% of the compartments and correspondence in the detection of lesions in 75.94% of them. The shown results assessed the clinical potential of this tool and present a step forward into structured MSK imaging reports.
|
|||
4049.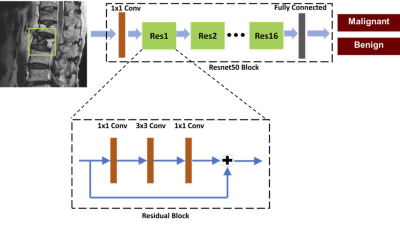 |
Differentiation of Benign and Malignant Vertebral Fractures on Spine MRI Using ResNet Deep Learning Compared to Radiologists’ Reading
Lee-Ren Yeh1, Yang Zhang2, Jeon-Hor Chen2, An-Chi Wang3, JieYu Yang3, Peter Chang2, Daniel Chow2, and Min-Ying Su2
1Radiology, E-Da Hospital, Kaohsiung, Taiwan, 2University of California Irvine, Irvine, CA, United States, 3Radiology, Chi-Mei Medical Center, Tainan, Taiwan
This study compared the reading of three radiologists with different level of experience, and also investigated the potential of deep learning to differentiate between benign and malignant vertebral fractures based on T1W and T2W MRI. The results showed that deep learning using ResNet50 achieved a satisfactory diagnostic accuracy of 92%, although inferior to 98% made by a senior MSK radiologist and 96% made by a R4 resident, much higher compared to 66% made by a R1 resident. The inferior performance of ResNet50 might be partly explained by the very limited information when only considering a small bounding box.
|
|||
4050.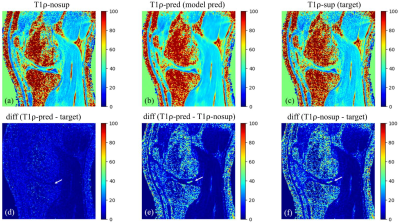 |
Synovial Fluid Suppressed 3D T1ρ Mapping of Knee Cartilage using Deep Learning
Can Wu1,2 and Qi Peng3
1Department of Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Philips Healthcare, Andover, MA, United States, 3Department of Radiology, Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, NY, United States
3D T1ρ mapping is a promising technique for quantitative assessment of biochemical changes in knee cartilage. However, synovial fluid, if not suppressed, may compromise T1ρ quantification, particularly in clinical conditions like osteoarthritis where cartilage is usually irregular and synovial fluid is increased. A long-T2-selective inversion approach can be used to suppress the synovial fluid signal at the cost of increased scan time by 50%. This study demonstrated that deep learning can be used to effectively eliminate synovial fluid from T1ρ data acquired without active fluid suppression, potentially leading to improved T1ρ quantification of knee cartilage accuracy without adding scan time.
|
|||
4051.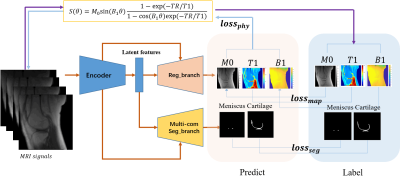 |
Deep CNNs with Physical Constraints for simultaneous Multi-tissue Segmentation and Quantification (MSQ-Net) of Knee from UTE MRIs
Xing Lu1, Yajun Ma1, Saeed Jerban1, Hyungseok Jang1, Yanping Xue1, Xiaodong Zhang1, Mei Wu1, Amilcare Gentili1,2, Chun-nan Hsu3, Eric Y Chang1,2, and Jiang Du1
1Department of Radiology, University of California, San Diego, San Diego, CA, United States, 2Radiology Service, Veterans Affairs San Diego Healthcare System, San Diego, CA, United States, 3Department of Neurosciences, University of California, San Diego, San Diego, CA, United States
In this study, we proposed end-to-end deep learning convolutional neural networks to perform simultaneous segmentation and quantification (MSQ-Net) on the knee without and with physical constraint networks (pcMSQ-Net). Both networks were trained and tested for the feasibility of simultaneous segmentation and quantitative evaluation of multiple knee joint tissues from 3D ultrashort echo time (UTE) magnetic resonance imaging. Results demonstrated the potential of MSQ-Net and pcMSQ-Net for fast and accurate UTE-MRI analysis of the knee, a “whole-organ” approach which is impossible with conventional clinical MRI.
|
|||
4052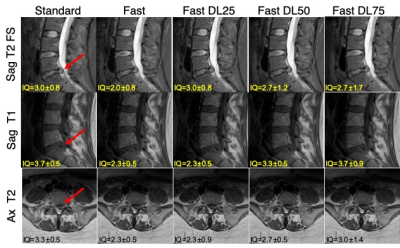 |
Deep-Learning Based Image Reconstruction for Lumbar Spine MRI at 3T: Clinical Feasibility Video Permission Withheld
Emma Bahroos1, Misung Han1, Cynthia Chin1, David Shin2, Javier Villanueva-Meyer1, Thomas Link1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2Applications and Workflow, GE Healthcare, Menlo Park, CA, United States
Lower back pain is one of the most common health problems, for which MRI is extensively used. Standard clinical, and fast acquisition images of lumbar spine were acquired for 18 patients. A (DL)-based image reconstruction was applied to the raw data of the fast images, with 25%, 50%, and 75% noise reduction factors. Evaluation of fast images with DL algorithm, for image quality, diagnostic capability, and SNR to standard images was conducted by three experienced radiologists. Our results show SNR improvement with higher noise reduction factor without a severe degradation in the ability to discern anatomical structures.
|
|||
4053.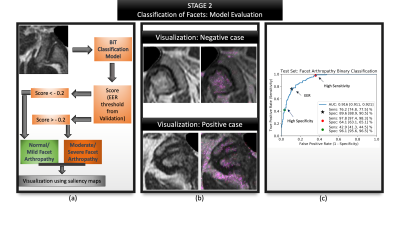 |
Less is more: zero-shot detection and transfer learning for facet arthropathy localization and classification on lumbar spine MRIs
Upasana Upadhyay Bharadwaj1, Cynthia T Chin1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology, University of California, San Francisco, San Francisco, CA, United States
Lumbar facet arthropathy is frequently observed along with other degenerative changes of the spine in patients presenting with chronic low back pain. Deep learning has demonstrated unprecedented success in automated assessment of many spine degenerative changes, but heretofore not applied to facet arthropathy. This study presents binary classification of facet arthropathy (normal/mild vs moderate/severe) on T2-weighted axial MRI slices using a two-staged approach: zero-shot facet detection followed by classification. Our model achieves an AUC of 0.916 [0.911, 0.921] with sensitivity and specificity of 97.8% [97.4, 98.3] and 64.1% [63.1, 65.1], respectively and can potentially enhance the clinical workflow.
|
|||
 |
4054.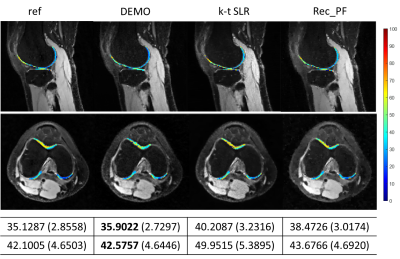 |
DEMO: Deep MR Parametric Mapping using Unsupervised Multi-tasking Framework
Jing Cheng1, Yuanyuan Liu1, Xin Liu1, Hairong Zheng1, Yanjie Zhu1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
In this work, we propose a novel deep learning-based framework DEMO for fast and robust MR parametric mapping. Different from current deep learning-based methods, DEMO trains the network in an unsupervised way. Specifically, a CS-based loss function is used in DEMO to avoid the necessity of using fully sampled k-space data as the label, and thus make it an unsupervised learning approach. DEMO reconstructs the parametric weighted images and generates the parametric map simultaneously, which enables multi-tasking learning. Experimental results show the promising performance of the proposed DEMO framework in quantitative MR T1ρ mapping.
|
||
4055.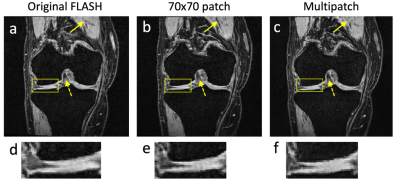 |
MRI image synthesis with a conditional generative adversarial network using patch pooling
Bragi Sveinsson1,2 and Matthew S Rosen1,2,3
1Martinos Center, Massachusetts General Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Physics, Harvard University, Cambridge, MA, United States
Deep learning networks allow the creation of new images based on a separate set of reference image data. This can be used to synthesize a specific MRI contrast from other image contrasts sharing the same anatomy. A particularly successful approach uses a conditional generative adversarial network with a patch-based discriminator, processing image patches of a fixed size. In this work, we investigate the benefits of using multiple patch sizes to improve image quality.
|
|||
4056.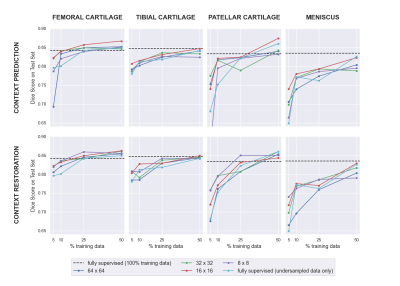 |
Self-Supervised Deep Learning for Knee MRI Segmentation using Limited Labeled Training Datasets
Jeffrey Dominic1, Arjun Desai1, Andrew Schmidt1, Elka Rubin1, Garry Gold1, Brian Hargreaves1, and Akshay Chaudhari1
1Stanford University, Stanford, CA, United States
Deep learning (DL)-based approaches have shown promise for automating medical image segmentation with high efficacy. However, current state-of-the-art DL supervised methods require large extents of labeled training images, which are difficult to curate at scale. In this work, we propose a self-supervised training scheme to reduce dependence on labeled data by pretraining networks in an unsupervised manner. We show that our method can improve segmentation performance, especially in the context of very limited data scenarios (only 10-25% scans available) and can achieve or surpass the accuracy of state-of-the-art supervised networks with approximately 50% fewer labeled scans.
|
|||
4057.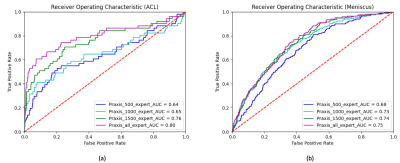 |
Deep Learning Improves Detection of Anterior Cruciate Ligament- and Meniscus Tear Detection in Knee MRI
Firas Khader1, Gustav Müller-Franzes1, Johannes Stegmaier2, Martin Pixberg3, Jonas Müller-Hübenthal3, Christiane Kuhl 1, Sven Nebelung4, and Daniel Truhn1
1Department of Diagnostic and Interventional Radiology, Aachen University Hospital, Aachen, Germany, 2Institute of Imaging and Computer Vision, RWTH Aachen University, Aachen, Germany, 3Praxis im Köln Triangle, Cologne, Germany, 4Department of Diagnostic and Interventional Radiology, Düsseldorf University Hospital, Dusseldorf, Germany
In this study we aimed to analyze the capability of neural networks to accurately diagnose the presence of ACL and meniscus tears in our in-house dataset comprised of 3887 manually annotated knee MRI exams. To this end we trained the MRNet architecture on a varying number of training exams that included proton density-weighted axial, sagittal and coronal planes for each knee exam. Additionally, we compared the performance of the architecture when trained on expert vs non-expert annotations. This study demonstrates that while our neural network benefits from a larger dataset, expert annotations do not considerably improve the performance.
|
|||
4058.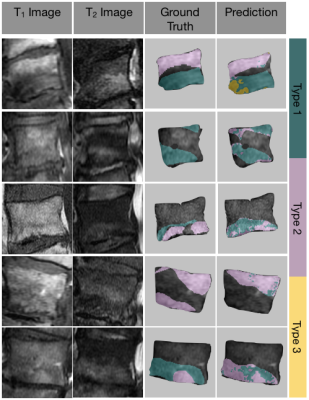 |
Fully automatic detection and voxel-wise mapping of vertebral body Modic changes using deep convolutional neural networks
Kenneth T Gao1,2,3, Radhika Tibrewala1,2, Madeline Hess1,2, Upasana Bharadwaj1,2, Gaurav Inamdar1,2, Cynthia T Chin1, Valentina Pedoia1,2, and Sharmila Majumdar1,2
1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Center for Intelligent Imaging, Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 3University of California, Berkeley-University of California San Francisco Graduate Program in Bioengineering, San Francisco, CA, United States
Modic changes are common degenerative lesions seen in spinal MRI and are strongly linked to lower back pain. However, detection of Modic changes suffers from poor inter-operator and inter-scanner reliabilities. We present a fully automatic, quantitative model that leverages deep learning and signal-based clustering for mapping Modic changes from clinically acquired MRI. The model achieves an identification rate of 85.7% and substantial agreement with radiologists. More importantly, the mapping technique classifies detected lesions on a voxel-wise basis, allowing for assessment of sensitive, local pathologies.
|
|||
4059.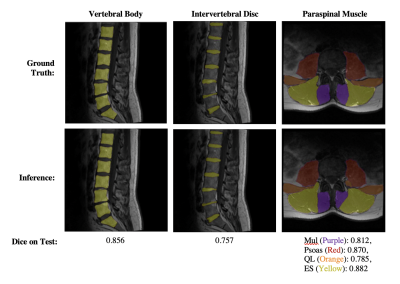 |
Towards Clinical Translation of Fully Automatic Segmentation and 3D Biomarker Extraction of Lumbar Spine MRI
Madeline Hess1, Kenneth Gao1, Radhika Tibrewala1, Gaurav Inamdar1, Upasana Bharadwaj1, Cynthia Chin1, Valentina Pedoia1, and Sharmila Majumdar1
1Center for Intelligent Imaging, University of California, San Francisco, San Francisco, CA, United States
Lumbar spine segmentation serves as an important first step for automated disease classification and monitoring, but manual segmentation is costly and time consuming. We present a deep learning-based pipeline to automatically segment the vertebral bodies, intervertebral discs, and paraspinal muscles in the lumbar spine. We leverage the results of this method to quickly and accurately extract disc height with a mean absolute error of 2.09 mm, muscle CSA with mean absolute errors of less than 1.46 cm2, and muscle centroid position with a mean absolute error of less than 7.23mm.
|
|||
 |
4060.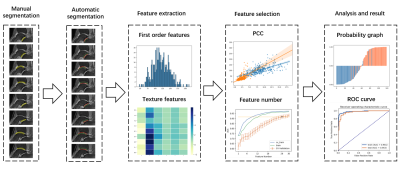 |
A pipeline combining deep learning and radiomics to automatically identify chronic lateral ankle instability from FS-PD MRI
Yibo Dan1, Hongyue Tao2, Chengxiu Zhang1, Chenglong Wang1, Yida Wang1, Shuang Chen2, and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, shanghai, China, 2Department of Radiology, Huashan Hospital, Fudan University, shanghai, China
The naked eye can only recognize the morphological changes of cartilage and subchondral bone on conventional MRI, but cannot recognize the subtle changes in their internal structure. The aim is to use radiomics to evaluate the cartilage and subchondral bone changes in patients with chronical ankle joint instability (CAI) on conventional MRI images1. We built a pipeline to automatically identify CAI from FS-PD images. The pipeline automatically segmented cartilage regions and subchondral bone (5mm) regions, then used SVM based on radiomics features extracted from these regions for classification. In the test dataset, the proposed model achieved an AUC of 0.965.
|
||
4061.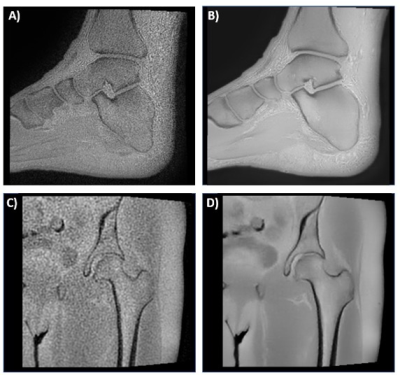 |
Deep Learning Reconstruction of 3D Zero Echo Time Magnetic Resonance Images for the Creation of 3D Printed Anatomic Models
Nicole Wake1,2, Stephanie Shamir1, Beverly Thornhill1, Nogah Haramati1, Graeme McKinnon3, Mathias Engstrom4, Florian Wiesinger4, Michael Carl5, Fraser Robb6, and Maggie Fung7
1Department of Radiology, Montefiore Medical Center, Bronx, NY, United States, 2Center for Advanced Imaging Innovation and Research, Department of Radiology, NYU Langone Health, New York, NY, United States, 3GE Healthcare, Waukesha, WI, United States, 4GE Healthcare, Munich, Germany, 5GE Healthcare, San Diego, CA, United States, 6GE Healthcare, Aurora, OH, United States, 7GE Healthcare, New York, NY, United States
Patient-specific three-dimensional (3D) printed anatomic models are valuable clinical tools which are generally created from computed tomography (CT). However, magnetic resonance imaging (MRI) is an attractive alternative, since it offers exquisite soft-tissue characterization and flexible image contrast mechanisms while avoiding the use of ionizing radiation. The purpose of this study was to evaluate the image quality and assess the feasibility of creating 3D printed models using a 3D Zero echo time (ZTE) MR images which were reconstructed with a deep learning reconstruction method.
|
|||
4062.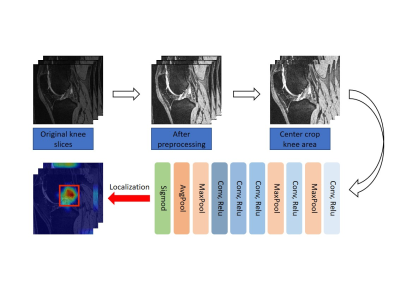 |
Identification of Bone Marrow Lesions on Magnetic Resonance Imaging with Weakly Supervised Deep Learning
Jiaping Hu1, Zhao Wang2, Lijie Zhong1, Keyan Yu1, Yanjun Chen1, Yingjie Mei3, Qi Dou4, and Xiaodong Zhang1
1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, Guangzhou, China, 2College of Information Science and Electronic Engineering, Zhejiang University, Hangzhou, China, 3China International Center, Philips Healthcare, Guangzhou, China, 4Department of Computer Science & Engineering, The Chinese University of Hong Kong, Hong Kong, China
The presence of a bone marrow lesion is associated with incident and progressive knee osteoarthritis (KOA) and joint replacement. Since the ill-defined boundary and various signal strength, identification of bone marrow lesions (BMLs) requires professional diagnostic ability and is subjective. Therefore, we utilize a model to assess whether there exists BMLs in every subregion and their severity on 3D-dual echo steady state (DESS) images according to MRI Osteoarthritis Knee Score (MOAKS). The initiatory results showed that deep learning framework performed well on discrimination of BMLs with good reproducibility.
|
|||
4063.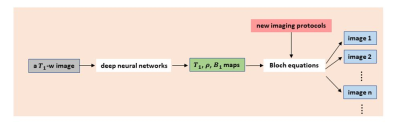 |
Retrospective Contrast Tuning from a Single T1-weighted Image Using Deep Learning
Yan Wu1, Yajun Ma2, Jiang Du2, and Lei Xing1
1Stanford University, Palo Alto, CA, United States, 2University of California San Diego, San Diego, CA, United States
While versatile soft tissue contrasts are achievable in MRI, contrast attainable from each scan is predetermined by the imaging protocol. A retrospective tuning of contrast will provide an opportunity to normalize MRI data for radiomics analysis. In this study, we present a new paradigm to obtain a spectrum of contrasts from a single T1-weighted image. Using deep learning, T1 map, proton density map, and B1 map are predicted from every T1-weighted image, and new contrasts can be obtained with the application of Bloch equations. The method has been validated in knee MRI with high accuracy achieved.
|
|||
4064.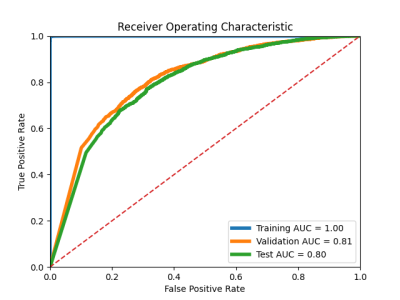 |
Feasibility of Femoral Cartilage Lesion Classification on Clinical MRIs using Deep Learning
Mingrui Yang1, Ceylan Colak1, Mercan Aslan1, Sibaji Gaj1, Morgan Jones1, Carl Winalski1, Naveen Subhas1, and Xiaojuan Li1
1Cleveland Clinic, Cleveland, OH, United States
Early diagnosis and effective detection of cartilage degeneration is an important factor for osteoarthritis prevention and treatment, which are still challenging in routine clinical practice, resulting in poor patient treatment and management plans. The purpose of this study is to assess the feasibility of building an automatic femoral cartilage lesion classification pipeline for heterogenous clinical routine MR scans by combining deep learning segmentation and classification models together.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.