Digital Posters
Emerging Applications of AI in Neuroimaging for CES I
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 6 | 19:00 - 20:00 |
 |
3481.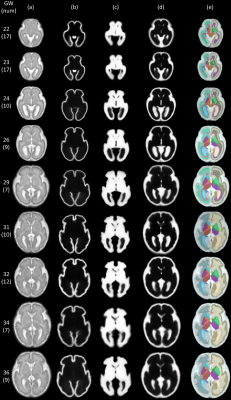 |
Learning 4D Probabilistic Atlas of Fetal Brain with Multi-channel Registration Network
Yuchen Pei1, Fenqiang Zhao1, Liangjun Chen1, Zhengwang Wu1, Tao Zhong1, Ya Wang1, Li Wang1, He Zhang2, and Gang Li1
1Department of Radiology and BRIC, University of North Carolina at Chapel Hill, USA, Chapel Hill, NC, United States, 2Department of Radiology, Obstetrics and Gynecology Hospital, Fudan University, Shanghai, China, Shanghai, China
Brain atlases are of fundamental importance for analyzing the dynamic neurodevelopment in fetal brains. Since the brain size, shape, and structure change rapidly during the prenatal development, it is essential to construct a spatiotemporal (4D) atlas with tissue probability maps for accurately characterizing dynamic changes in fetal brains and providing tissue prior for segmentation of fetal brain MR images. We propose a novel unsupervised learning framework for building multi-channel atlases by incorporating tissue segmentation. Based on 98 healthy fetuses from 22 to 36 weeks, the learned 4D fetal brain atlas includes intensity templates, corresponding tissue probability maps and parcellation maps.
|
||
3482.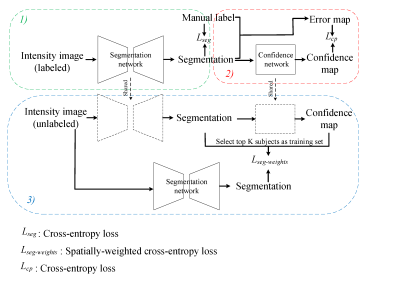 |
Self-Supervised Transfer Learning for Infant Cerebellum Segmentation with Multi-Domain MRIs
Yue Sun1, Kun Gao1, Shihui Ying1, Weili Lin1, Gang Li1, Sijie Niu1, Mingxia Liu1, and Li Wang1
1Department of Radiology and Biomedical Research Imaging Center, UNC at Chapel Hill, Chapel Hill, NC, United States
This study develops a self-supervised transfer learning (SSTL) framework to generate reliable cerebellum segmentations for infant subjects with multi-domain MRIs, aiming to alleviate the domain shift between different time-points/sites and improve the generalization ability. Experiments demonstrate that by transferring limited manual labels from late time-points (or a specific site) with high tissue contrast to early time-points (or other sites) with low contrast, our method achieves improved performance and can be applied to other tasks, especially for those with multi-site data.
|
|||
3483.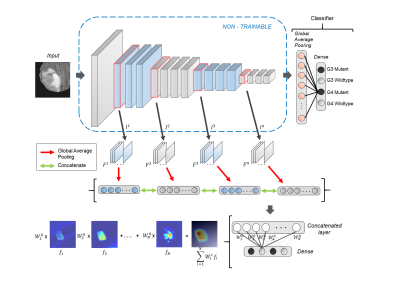 |
MRI based IDH and grade prediction using convolutional neural networks
Sumeet Shinde1, Abhilasha Indoria2, Jitender Saini2, Manish Beniwal2, Vani Santosh2, and Madhura Ingalhalikar1
1Symbiosis centre for medical image analysis, Symbiosis International University, Pune, India, 2Dept of Radiology, National Institute of Mental Health and Neurosciences, Bengaluru, India
Recent developments in glioma subtyping suggest that IDH genotype as well as the histological grading are both crucial factors. However, earlier classification studies based on MRI features have focused either only on grade or IDH. In this work we employ an automated deep learning based technique to delineate the grade as well as the IDH status on a dataset of 178 subjects. Our classifier performs with a superior accuracy of 93.5% and the model explanability is achieved through class activation maps that illustrate the areas important in the classification.
|
|||
3484.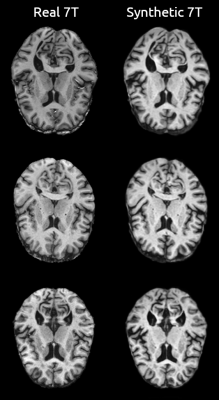 |
Image-to-image translation of 3T to 7T MRI using Generative Adversarial Networks: A step towards longitudinal harmonization
Eduardo Diniz1, Karim Helmet2, Tales Santini2, Howard Eizenstein2, and Tamer Ibrahim2
1Electrical and Computer Engineering, University of Pittsburgh, Pittsburgh, PA, United States, 2Bioengineering, University of Pittsburgh, Pittsburgh, PA, United States
We implemented a generative adversarial network (CycleGAN) to tackle the problem of MRI data harmonization across scanner strength. We leveraged a large dataset of unpaired 3T and 7T MR images for training and evaluated our model in a dataset of paired 3T and 7T data by generating synthetic 7T images and comparing them with their real counterparts. Dice scores and volumetric measures showed strong agreement between the synthetic and real 7T images. This approach allows for research studies to transition from 3T to 7T MR systems, thereby harnessing 7T systems advantages without losing the prior wave’s 3T MR data.
|
|||
3485.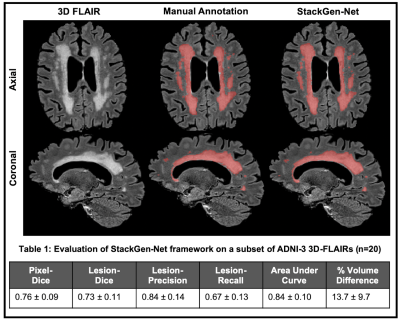 |
White matter hyperintensity volumes and cognition: Assessment of a deep learning-based lesion detection and quantification algorithm on ADNI
Lavanya Umapathy1, Gloria Guzman Perez-Carillo2, Blair Winegar3, Srinivasan Vedantham4, Maria Altbach4, and Ali Bilgin1,4,5
1Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2Mallinckrodt Institute of Radiology, St Louis, MO, United States, 3Radiology and Imaging Sciences, University of Utah, Salt Lake, UT, United States, 4Medical Imaging, University of Arizona, Tucson, AZ, United States, 5Biomedical Engineering, University of Arizona, Tucson, AZ, United States
The relationship between cognition and white matter hyperintensities (WMH) volumes often depends on accuracy of the lesion segmentation algorithm used. As such, accurate detection and quantification of WMH is of great interest. Here, we use a deep learning-based WMH segmentation algorithm, StackGen-Net, to detect and quantify WMH on 3D-FLAIR images from ADNI. We used a subset of subjects (n=20) and obtained manual WMH segmentations by an experienced neuro-radiologist to demonstrate the accuracy of our algorithm. On a larger cohort of subjects (n=290), we observed larger WMH volumes correlated with worse performance on executive function (P=.004), memory (P=.01), and language (P=.005).
|
|||
3486.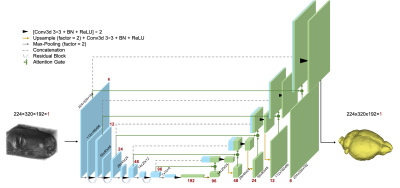 |
DL-BET - A deep learning based tool for automatic brain extraction from structural magnetic resonance images in mice.
Sabrina Gjerswold-Selleck1, Nanyan Zhu2,3,4, Haoran Sun1, Dipika Sikka1, Jie Shi1, Chen Liu3,4,5, Tal Nuriel4,6, Scott A. Small4,7,8, and Jia Guo3,9
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Biological Science, Columbia University, New York, NY, United States, 3Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States, 4Taub Institute for Research on Alzheimer’s Disease and the Aging Brain, Columbia University, New York, NY, United States, 5Electrical Engineering, Columbia University, New York, NY, United States, 6Department of Pathology and Cell Biology, Columbia University, New York, NY, United States, 7Gertrude H. Sergievsky Center, Columbia University, New York, NY, United States, 8Radiology, Columbia University, New York, NY, United States, 9Psychiatry, Columbia University, New York, NY, United States
Brain extraction plays an integral role in image processing pipelines in both human and small animal preclinical MRI studies. Due to lack of state-of-the-art tools for automated brain extraction in rodent research, this step is often performed semi-supervised with manual correction, making it prone to inconsistent results. Here, we perform a multi-model brain extraction study and present a semi-automated preprocessing workflow and deep neural network with a 3D Residual Attention U-Net architecture as the optimal network for automated skull-stripping in neuroimaging analysis pipelines, achieving a DICE score of 0.987 and accuracy of 99.7%.
|
|||
3487.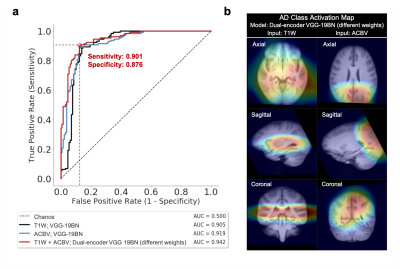 |
Deep Learning Identifies Neuroimaging Signatures of Alzheimer’s Disease Using Structural and Artificial Functional MRI Data
Nanyan Zhu1,2,3, Chen Liu2,3,4, Sabrina Gjerswold-Selleck5, Xinyang Feng5, Dipika Sikka5, Scott A. Small2,6,7, and Jia Guo3,8
1Biological Science, Columbia University, New York, NY, United States, 2Taub Institute for Research on Alzheimer's Disease and the Aging Brain, Columbia University, New York, NY, United States, 3Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States, 4Electrical Engineering, Columbia University, New York, NY, United States, 5Biomedical Engineering, Columbia University, New York, NY, United States, 6Radiology, Columbia University, New York, NY, United States, 7Gertrude H. Sergievsky Center, Columbia University, New York, NY, United States, 8Psychiatry, Columbia University, New York, NY, United States
Alzheimer’s disease (AD) is a neurodegenerative disorder where functional deficits precede structural deformations. Various studies have demonstrated the efficacy of deep learning in diagnosing AD using imaging data, and that functional modalities are more helpful than structural counterparts over comparable sample size. To deal with the lack of large-scale functional data in the real world, we used a structure-to-function translation network to artificially generate a previously non-existent spatially-matched functional neuroimaging dataset from existing large-scale structural data. The artificial functional data, generated with little cost, complemented the authentic structural data to further improve the performance of AD classification.
|
|||
3488.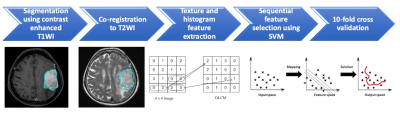 |
Preoperative MR Radiomics and ADC Value for Prediction of Progression and Recurrence in Meningiomas
Ching-Chung Ko1,2, Yang Zhang3, Jeon-Hor Chen3,4, and Min-Ying Su3
1Department of Medical Imaging, Chi Mei Medical Center, Tainan, Taiwan, 2Department of Health and Nutrition, Chia Nan University of Pharmacy and Science, Tainan, Taiwan, 3Department of Radiological Sciences, University of California, Irvine, CA, CA, United States, 4Department of Radiology, E-Da Hospital and I-Shou University, Kaohsiung, Taiwan
A subset of meningiomas may show early progression/recurrence (P/R) after surgery. In clinical practice, one of the main challenges in the treatment of meningiomas is to determine factors that correlate with P/R. This study investigated the role of preoperative MR radiomics and apparent diffusion coefficient (ADC) value for prediction of P/R in meningiomas. The four most significant radiomic features selected by support vector machine (SVM) were used to calculate SVM score for each patient. High SVM score and low ADC value were associated with early P/R and short progression-free survival in meningiomas.
|
|||
3489.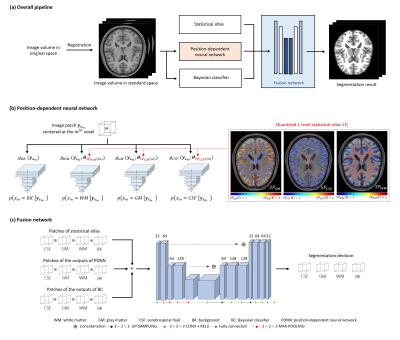 |
Improved Segmentation of MR Brain Images by Integrating Bayesian and Deep Learning-Based Classification
Ruihao Liu1, Ziyu Meng1, Wenli Li1, Yao Li1, Yiping P. Du1, and Zhi-Pei Liang2,3
1Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Accurate segmentation of brain tissues is essential for brain imaging applications. Classical Bayesian methods rely on good probability functions to produce good segmentation results. This paper presents a new method to synergistically integrate classical Bayesian segmentation with deep learning-based classification. A cluster of patch-based position-dependent neural networks were trained to effectively capture the joint spatial-intensity distributions of brain tissues. This cluster of patch networks significantly extends the capability of classical Markov Random field models and conventional statistical brain atlases. By combining the classical Bayesian classifier with the proposed networks, our method significantly improved segmentation results compared with the state-of-art methods.
|
|||
3490. |
Simulation of brain deficits on MRI: A novel approach of ‘ground truth’ generation for machine learning
Kattie Sepehri1, Xiaowei Song2, Ryan Proulx3, Sujoy Ghosh Hajra4, Brennen Dobberthien5, Careesa Liu6, Ryan D'Arcy7, Don Murray3, and Andra Krauze5
1UBC, Vancouver, BC, Canada, 2Surrey Memorial Hospital, Vancouver, BC, Canada, 3Safe Software, Vancouver, BC, Canada, 4National Research Council, Vancouver, BC, Canada, 5BC Cancer, Vancouver, BC, Canada, 6Baycrest Health Sciences Centre, Vancouver, BC, Canada, 7HealthTech Connex, Vancouver, BC, Canada
Robust machine learning algorithms for tumor identification require ground truth data sets. Ground truth data sets require expert input, are difficult and inefficient to produce. Feature Manipulation Engine (FME) allows for specific and complex data manipulation. We have created an FME workflow to produce simulated tumors that resemble realistic gliomas as rated by experts.
|
|||
3491.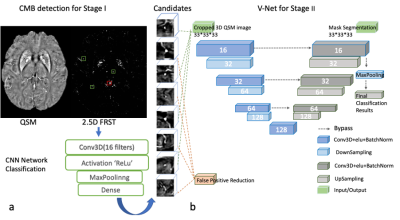 |
Deep learning-based Cerebral Microbleeds detection on quantitative susceptibility mapping(QSM) for Stroke Cohort
Peng Xia1, Zuojun Wang1, Han Yu1, Fan Huang1, Henry Ka-Fung MAK1, Edward Sai-am HUI2, and Peng Cao1
1Department of Diagnostic Radiology, The University of Hong Kong, Hong Kong, China, 2Department of Rehabilitation Science, The Hong Kong Polytechnic University, Hong Kong, China
Cerebral microbleeds (CMBs) detection has been applied to many brain diseases but not well studied on stroke cohort. We developed and analyzed a deep learning-based CMB detection pipeline on quantitative susceptibility mapping (QSM) images from 138 stroke patients cohort.
|
|||
3492.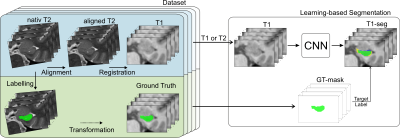 |
Improved Automated Hippocampus Segmentation using Deep Neural Networks
Maximilian Sackl1, Alina Dima2, Christian Payer2, Darko Štern3, Reinhold Schmidt1, and Stefan Ropele1
1Department of Neurology, Medical University of Graz, Graz, Austria, 2Institute of Computer Graphics and Vision, Graz University of Technology, Graz, Austria, 3Department of Biophysics, Medical University of Graz, Graz, Austria
Segmentation of the hippocampal formation on T1-weighted structural MR scans is a prerequisite for most imaging studies in Alzheimer’s disease. In this work, we evaluated the performance and accuracy of deep learning-based hippocampus segmentation combined with manual ground truth (GT) data that originates from high-resolution T2-weighted MR images. Results were evaluated against the GT-labels and compared to segmentation results obtained with FreeSurfer. All learning approaches outperformed FreeSurfer in terms of accuracy and speed, where experiments utilizing the T2-based GT-labels yielded the best results. Thus, using T2-weighted images for training a deep learning model can improve automated HC segmentation.
|
|||
3493.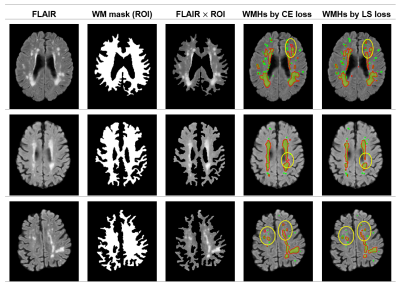 |
A semi-supervised level-set loss for white matter hyperintensities segmentation on FLAIR without manual labels
Fan Huang1, Peng Xia1, Varut Vardhanabhuti1, Edward Sai-Kam Hui2, Gary Kui-Kai Lau3, Henry Ka-Fung Mak1, and Peng Cao1
1Department of Diagnostic Radiology, The University of Hong Kong, Hong Kong, Hong Kong, 2Department of Rehabilitation Science, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 3Department of Medicine, The University of Hong Kong, HongKong, Hong Kong
We propose a semi-supervised training scheme for white matter hyperintensity (WMHs) segmentation using V-Net on FLAIR images. The training procedure does not require manual labeling data but only a few domain knowledge of WMHs. The segmentation result obtained by the V-Net with the proposed scheme outperformed that obtained by the supervised loss with manual labels, showing great potential and generalizability in medical image applications.
|
|||
3494.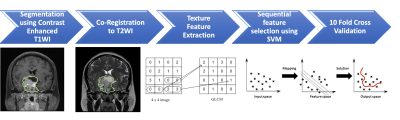 |
Preoperative MR Radiomics for Prediction of Progression and Recurrence in Non-functional Pituitary Macroadenomas
Ching-Chung Ko1,2, Yang Zhang3, Jeon-Hor Chen3,4, and Min-Ying Su3
1Department of Medical Imaging, Chi Mei Medical Center, Tainan, Taiwan, 2Department of Health and Nutrition, Chia Nan University of Pharmacy and Science, Tainan, Taiwan, 3Department of Radiological Sciences, University of California, Irvine, CA, CA, United States, 4Department of Radiology, E-Da Hospital and I-Shou University, Kaohsiung, Taiwan
A subset of nonfunctioning pituitary macroadenomas (NFPAs) may show early progression/recurrence (P/R) after surgery. In clinical practice, one of the main challenges in the treatment of NFPAs is to determine factors associated with P/R. This study investigated the role of preoperative MR radiomics based on support vector machine (SVM) for the prediction of P/R in NFPAs. The three most significant radiomic features selected by SVM were used to calculate SVM score for each patient. High SVM scores were associated with P/R and short progression-free survival in NFPAs.
|
|||
3495.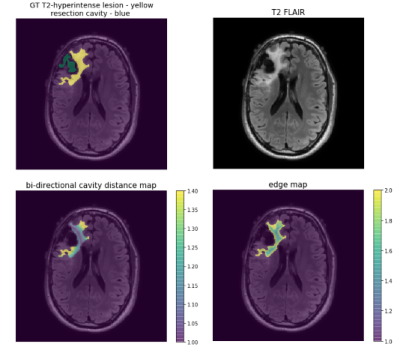 |
Improving the generalizability of convolutional neural networks for T2-lesion segmentation of gliomas in the post-treatment setting
Jacob Ellison1, Francesco Caliva1, Pablo Damasceno2, Tracy Luks1, Marisa LaFontaine1, Julia Cluceru1, Anil Kemisetti1, Yan Li1, Valentina Pedoia2, Javier Villanueva-Meyer1, and Janine M Lupo1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Center for Intelligent Imaging, UCSF, San Francisco, CA, United States
Routine monitoring of response to therapy in patients with glioma greatly benefits from using volumetrics quantified from lesion segmentation. Yet, the vast majority of deep learning models developed for this task have been trained using data from treatment-naïve, newly-diagnosed patients, whose T2-lesions have different appearance on imaging. We found that increasing the proportion of treated patients in training, incorporating a cross-entropy loss term that takes into account the spatial distance from surgical resection cavity and leading tumor edge, and transfer learning from newly-diagnosed to post-treatment imaging domains were effective strategies to improve the generalizability of segmentation of the T2-lesion post-treatment.
|
|||
3496.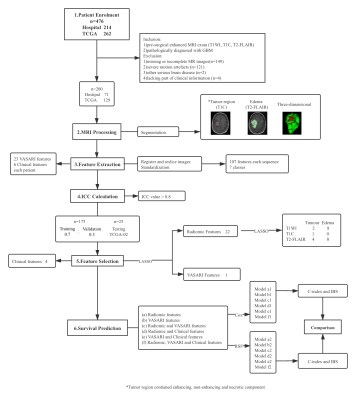 |
A Comparative and Summative Study of Radiomics-Based Overall Survival Prediction in Glioblastoma Patients
Zhuoying Ruan1, Nan Mei2, Yiping Lu2, Ji Xiong3, Xuanxuan Li2, Yajing Zhao2, Pu-Yeh Wu4, Li Liu5, and Bo Yin2
1Shanghai Institute of Medical Imaging, Fudan University, Shanghai, China, 2Department of Radiology, Huashan Hospital Fudan University, Shanghai, China, 3Department of Pathology, Huashan Hospital Fudan University, Shanghai, China, 4GE Healthcare, MR Research China, Shanghai, China, 5Department of Radiology, Shanghai Cancer Center, Fudan University, Shanghai, China The prediction of overall survival in glioblastoma patients can provide great aid to clinical treatment. Therefore, we extracted clinical, VASARI and radiomic features from pre-operative MRI scannings and compared the predictive capacity of Cox algorithm and random forest based on these features. According to our results, tumor without cortical involvement had longer overall survival than those involved; the random forest models outperformed Cox regression models in general and the random forest model consisting of radiomic features was the best one. Ten radiomic features were reproducible in our and other’s studies exhibiting promising value in overall survival prediction of glioblastoma. |
|||
3497.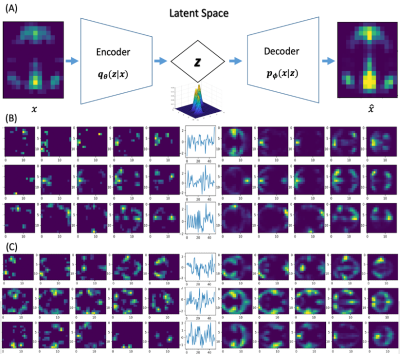 |
Mapping transient coactivity patterns of brain in latent space with variational autoencoder neural network
Kaiming Li1 and Xiaoping Hu1
1Department of Bioengineering, UC Riverside, Riverside, CA, United States
The brain is a complex dynamic system that constantly evolves. Characterization of the spatiotemporal dynamics of brain activity is fundamental to understand how brain works. Current studies with functional connectivity and linear models are limited by sacrificed temporal resolution and insufficient model capacity. With a generative variational auto encoder (VAE), the present study mapped the high-dimensional transient co-activity patterns (CAPs) of large datasets in a low-dimensional latent space. We demonstrated with multiple datasets that VAE can effectively represent the transient CAPs in latent space, paving the way for frame-wise modeling of the complex spatiotemporal dynamics in future.
|
|||
3498.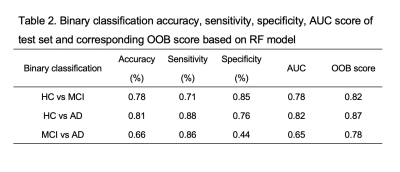 |
Classification of Alzheimer's Disease Based on Amyloid-PET using Random Forest Ensemble
Yiwen Bao1, Patrick Ka-Chun Chiu2, Yat-Fung Shea2, Joseph SK Kwan3, Felix Hon Wai Chan2, and Henry Ka-Fung Mak1
1Department of diagnostic radiology, University of Hong Kong, Hong Kong, Hong Kong, 2Department of medicine, Queen Mary Hospital, Hong Kong, Hong Kong, 3Department of brain sciences, Imperial College London, London, United Kingdom
Random forest model as a high efficacy classifier was incorporated in our study for supporting clinical diagnosis. We aimed at evaluating the accuracy of RF model in distinguishing HC, MCI from AD and the importance of various neuroradiological features in selection. Additionally, in order to unify quantitative amyloid uptake across three cohorts, we transformed SUVR into standard Centiloid unit. The results indicated that RF model had moderate to high accuracy in differentiating AD from HC and MCI. Regional Ab load had more important effects than other features in distinguishing AD from others.
|
|||
3499.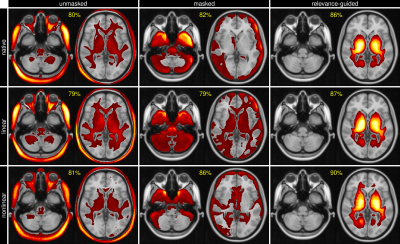 |
Increasing Feature Sparsity in Alzheimer's Disease Classification with Relevance-Guided Deep Learning
Christian Tinauer1, Stefan Heber1, Lukas Pirpamer1, Anna Damulina1, Reinhold Schmidt1, Stefan Ropele1, and Christian Langkammer1
1Department of Neurology, Medical University of Graz, Graz, Austria
Using T1-weighted images we separated Alzheimer's patients (n=130) from healthy controls (n=375) by using a deep neural network and found that the preprocessing steps might introduce unwanted features to be used by the classifier. We systematically investigated the influence of registration and brain extraction on the learned features using a relevance map generator attached to the classification network. The results were compared to our relevance-guided training method. Relevance-guided training identifies sparser but substantially more relevant voxels, which improves the classification accuracy.
|
|||
3500.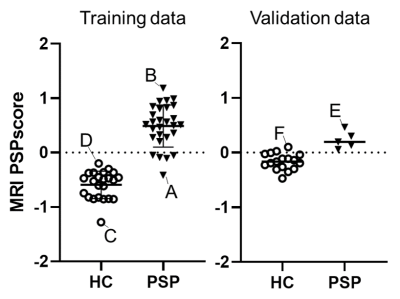 |
Establishment of the automated diagnostic system using MR imaging for patients with progressive supranuclear palsy based on in vivo tau imaging
Hironobu Endo1, Yuhei Takado1, Kenji Tagai1, Matsuoka Kiwamu1, Manabu Kubota2, Yasunori Sano1, Keisuke Takahata1, Maiko Ono1, Chie Seki1, Hideki Matsumoto1,3, Oya Masaki1, Yoko Ikoma1, Kazunori Kawamura1, Ming-rong Zhang1,
Hitoshi Shinotoh1,4, Kenichi Oishi5, Susumu Mori5, Takahiko Tokuda1, Hitoshi Shimada1, and Makoto Higuchi1
1National Institute of Radiological Sciences, National Institutes for Quantum and Radiological Science and Technology, Chiba, Japan, 2Department of Psychiatry, Kyoto University Graduate School of Medicine, Kyoto, Japan, 3Department of Oral and Maxillofacial Radiology, Tokyo Dental College, Tokyo, Japan, 4Neurology Clinic Chiba, Chiba, Japan, 5Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States
MRI-based diagnostic marker is desired for recruiting patients with progressive supranuclear palsy (PSP) for clinical trials of tau-targeting therapies. Elastic Net analysis was applied to a set of 144 MRI-VOIs (MR images-volumes of interest) to determine the VOIs useful for discriminating tau-positive PSP from healthy controls. The MRI-PSP score, calculated from the analysis, demonstrated 90.0% and 91.1% accuracy for training and validation data. The PSP rating scale correlated well with the MRI-PSP score. The MRI-PSP score calculated by an unbiased analysis system would be a promising diagnostic marker and can potentially predict disease severity in PSP.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.