Digital Posters
New Frontiers of AI in Neuroimaging
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 6 | 17:00 - 18:00 |
3245.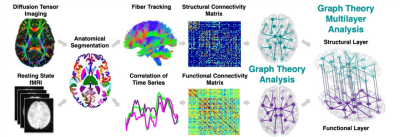 |
Classification Between Epilepsy Patients and Healthy Controls Using Multi-Modal Structure-Function Brain Network
Yael Jacob1, Gaurav Verma1, Lara Marcuse1, Madeline Fields1, and Priti Balchandani1
1Icahn School of Medicine at Mount Sinai, New York, NY, United States
Epilepsy patients (EP) endure harmful effects both on their health and quality of life. Early identification of these individuals would be incredibly helpful to gauge management and expectations. Implementing a novel multilayer network analysis, considering communication within functional and structural networks as well as the interactions between them, we tested whether this whole-brain comprehensive network hierarchy can be used as predictors of epilepsy. Using multilayer network features as predictors in a machine learning algorithm we were able to classify EP and controls with overall accuracy of 84%, demonstrating the applicability of multi-modal imaging for diagnostics of epilepsy.
|
|||
3246. |
No-Reference Quality Assessment of MRIs for Clinical Application
Ke Lei1, Shreyas Vasanawala2, and John Pauly1
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
We proposed a CNN model that automatically assesses image quality within seconds after a scan is finished to reduce the number of patient recalls and inadequate images. Our model is deployed to the clinics where it alerts technicians to take action for low-quality images while the patient is still in the scanner. Our model achieves super-human performance on assessing perceptual noise level in 2D fast spin echo (FSE) MRIs. It can also be used to automatically guide other computational processes, like training of a denoising model or choice of a regularization weight for reconstruction.
|
|||
3247.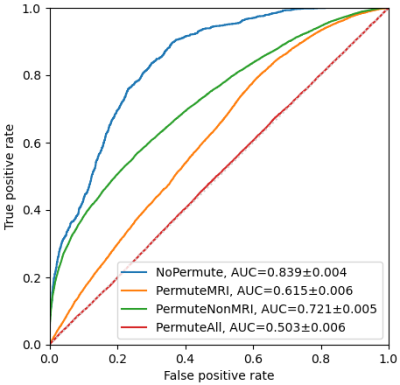 |
Importance of Clinical MRI Features in Predicting Epilepsy Drug Treatment Outcome for Pediatric Tuberous Sclerosis Complex
Jun Yang1,2, Cailei Zhao3, Shi Su4, Zhanqi Hu5, Jianxiang Liao5, Dong Liang1,2,4, and Haifeng Wang2,4
1Research Centre for Medical AI, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Department of Radiology, Shenzhen Children’s Hospital, Shenzhen, China, 4Paul C. Lauterbur Research Centre for Biomedical Imaging, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 5Department of Neurology, Shenzhen Children’s Hospital, Shenzhen, China
Predicting epilepsy drug treatment outcome is important for treating children with tuberous sclerosis complex (TSC). Here, the best performing model was selected to explore the contribution of the features, using permutation importance (PIMP). An approach similar to PIMP was used to compare the magnetic resonance imaging (MRI) and non-MRI features. The best model multilayer perceptron (MLP) with a hidden layer size of 60 and 30 features selected by F-test achieved the best performance. The results based on 103 children patients showed that some features were more important than others, and MRI features contributed more than non-MRI features in prediction.
|
|||
3248.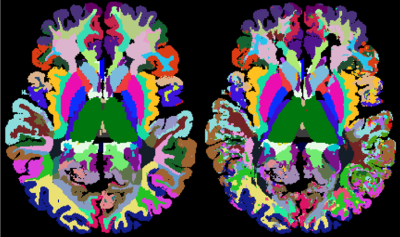 |
Brain tissues have single-voxel signatures in multi-spectral MRI
Alexander German1, Angelika Mennecke1, Jan Martin2, Jannis Hanspach1, Andrzej Liebert1, Jürgen Herrler1, Tristan Anselm Kuder3, Manuel Schmidt1, Armin Nagel1, Michael Uder1, Arnd Dörfler1, Jürgen Winkler1, Moritz Zaiss1,4,
and Frederik Laun1
1University Hospital Erlangen, Erlangen, Germany, 2Lund University, Lund, Sweden, 3German Cancer Research Center, Heidelberg, Germany, 4Max Planck Institute for Biological Cybernetics, Tübingen, Germany
We acquired diffusion-weighted and CEST data of the brain of 38 healthy volunteers. A MPRAGE and SWI based segmentation into 102 brain regions revealed unique diffusion and chemical MR signals on average. More importantly, we could infer these tissue classes form individual voxel data using a neural network. The revival of this old paradigm for tissue characterization from the 1990s points to the fact that unique MR signals of different brain regions exist and can be used to determine the tissue type voxel-wise. The approach as such is general and could unify the ever-growing diversity of MR contrasts.
|
|||
3249.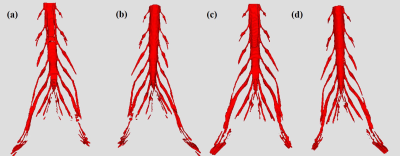 |
Deep Learning Approach for Lumbosacral Plexus Segmentation from Magnetic Resonance Neurography: Initial Study
Jian Wang1, Guohui Ruan2,3, Yingjie Mei4, Yanjun Chen1, Jialing Chen1, Yanqiu Feng2,3, and Xiaodong Zhang1
1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, Guangzhou, China, 2Guangdong Provincial Key Laboratory of Medical Image Processing, Southern Medical University, Guangzhou, China, 3School of Biomedical Engineering, Southern Medical University, Guangzhou, China, 4China International Center, Philips Healthcare, Guangzhou, China
Accurate regional segmentation of the Lumbosacral Plexus (LSP) on magnetic resonance neurography (MRN) images is a fundamental requirement before LSP related disorders diagnosis can be achieved. In this paper, we utilize U-Net to segment LSP trunk and branch from three-dimensional fast field echo(3D-FFE) with principle of selective excitation technique (Proset) images. The results show that a U-Net deep learning framework expresses highly performance and less time-consumption for LSP segmentation in patients with degenerative spinal diseases and healthy subjects.
|
|||
3250.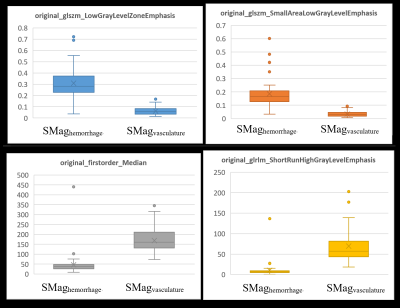 |
Differentiating hemorrhage and vasculature ITSS in SWI-magnitude images in intracranial Glioma: machine-learning and radiomic based approach
Rupsa Bhattacharjee1,2, Rakesh Kumar Gupta3, Suhail P Parvaze4, Rana Patir5, Sandeep Vaishya5, Sunita Ahlawat6, and Anup Singh1,7
1Center for Biomedical Engineering, Indian Institute of Technology (IIT) Delhi, New Delhi, India, 2Philips Health Systems, Philips India Limited, Gurugram, India, 3Department of Radiology, Fortis Memorial Research Institute, Gurugram, India, 4Philips Health Systems, Philips Innovation Campus, Bangalore, India, 5Department of Neurosurgery, Fortis Memorial Research Institute, Gurugram, India, 6SRL Diagnostics, Gurugram, India, 7Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, India
Intra-tumoral-susceptibility-signal (ITSS) has been increasingly proven to play a major role in glioma grading, progression assessment and follow-up. Quantitative ITSS assessment involves segmentation of ITSS from SWI images, separating vasculature ITSS from hemorrhage ITSS and finally quantifying the ITSS-vasculature-volume (IVV) to grade the glioma non-invasively. This study involves radiomic feature extraction, random-forest based feature selection and classification to indicate that radiomic features can significantly differentiate between 3Dvasculature and 3DHemorrhage mask regions in SWI-magnitude images. This is also one of the first studies that explores the vasculature and hemorrhage radiomic properties extracted from SWI-magnitude images through machine-learning in grade-IV GBM patients.
|
|||
3251.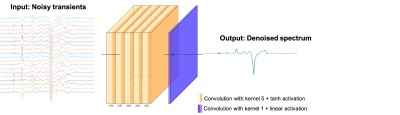 |
Reduction of J-difference Edited Magnetic Resonance Spectroscopy Acquisition Times Using Deep Learning
Roberto Souza1,2, Jordan McEwen3, Carissa Chung3, and Ashley D. Harris2,4
1Electrical and Computer Engineering, University of Calgary, Calgary, AB, Canada, 2Hotchkiss Brain Institute, Calgary, AB, Canada, 3Biomedical Engineering, University of Calgary, Calgary, AB, Canada, 4Radiology, University of Calgary, Calgary, AB, Canada
Magnetic Resonance Spectroscopy (MRS) non-invasively acquires in-vivo data on the chemical composition of localized tissue samples. MRS acquisitions are lengthy because they often require the acquisition of several averages to obtain a spectrum with a sufficient signal to noise ratio (SNR). This issue is augmented in J-difference edited MRS in which the analyzed spectrum is generated as the difference between sub-spectra in which editing pulses have been applied to selectively refocus the coupling of the target metabolite. In this work, we investigate the reduction of J-difference edited MRS acquisition times using deep learning.
|
|||
3252.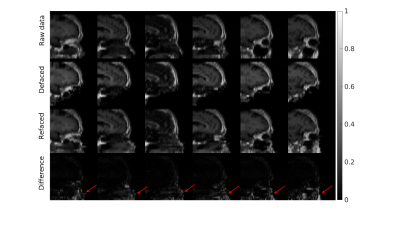 |
Defacing and Refacing Brain MRI Using a Cycle Generative Adversarial Network
Zuojun Wang1, Peng Xia1, Wenming Cao2, Kui Kai, Gary Lau1, Henry Ka Fung Mak3, and Peng Cao1
1Diagnostic Radiology, Department of Diagnostic Radiology, HKU, Hong Kong, China, 2Department of Diagnostic Radiology, HKU, HongKong, China, 3Department of Diagnostic Radiology, HKU, Hong Kong, China
MRI anonymizations, including face removal, are necessary for clinical data archiving and sharing. Segmentation based methods have been developed for semi-automated face removal on brain MRI. Meanwhile, the conventional methods are inefficient and unreliable, as the images have to be pre-processed and fed in the software manually. Deep learning-based methods are highly efficient in image-to-image translation on large scale databases. In this study, we utilized a cycle generative adversarial network to anonymize brain MRI data. The model showed reliable performance when testing on T1-weighted images, and we also extend it to the unseen MPRAGE images, targeting different brain MRI contrasts.
|
|||
3253.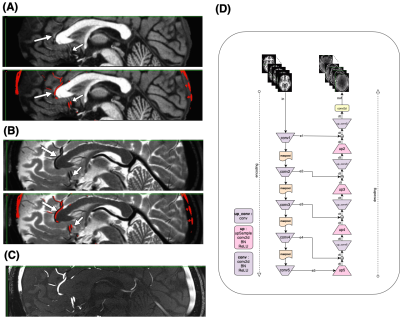 |
Deep Image Synthesis for Extraction of Vascular and Gray Matter Metrics
Farnaz Orooji1, Xinyang Wang1, Mohammed Ayoub Alaoui Mhamdi1, and Russell Butler1
1Computer Science, Bishop's University, Sherbrooke, QC, Canada
One of the main strengths of MRI is the wide range of soft tissue contrasts which can be obtained using different sequence parameters. T1-weighted-images provide exquisite contrast between gray/white matter but fail to capture arterial vessels. Here we experiment with deep-learning for image synthesis, to synthesize a time-of-flight-(TOF) angiogram based on T1 contrast image. We then compare arterial diameters from synthesized TOF with ground-truth-TOF diameters. We also synthesize a T1 from a T2, and compare cortical metrics such as thickness, curvature etc. We show that it is possible to obtain vessel diameters from T1, and cortical thick/vol/curv measures from T2.
|
|||
3254.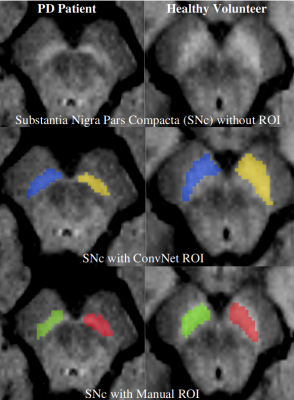 |
Substantia Nigra Abnormalities in Early Parkinson’s Disease Patients using Convolutional Neural Networks in Neuromelanin MRI
Rahul Gaurav1,2,3, Romain Valabregue1,2, Nadya Pyatigorskaya1,2,3,4, Lydia Yahia-Cherif1,2, Emma Biondetti1,2,3, Graziella Mangone2,5, R. Matthew Hutchison6, Jean-Christophe Corvol2,5,7, Marie Vidailhet2,3,7, and Stephane Lehericy1,2,3,4
1CENIR, ICM Paris, Paris, France, 2Paris Brain Institute (ICM), Sorbonne University, UPMC Univ Paris 06, Inserm U1127, CNRS UMR 7225, Paris, France, 3ICM Team “Movement Investigations and Therapeutics” (MOV’IT), Paris, France, 4Department of Neuroradiology, Pitié-Salpêtrière Hospital, AP-HP, Paris, France, 5INSERM, Clinical Investigation Center for Neurosciences, Pitié-Salpêtrière Hospital, Paris, France, 6Biogen Inc., Cambridge, MA, United States, 7Department of Neurology, APHP, Pitié-Salpêtrière Hospital, Paris, France
There is a need of accurate imaging biomarkers of dopaminergic cell neurodegeneration to facilitate drug trials in Parkinson’s disease (PD). PD demonstrates neurodegenerative substantia nigra pars compacta (SNc) changes that can be detected efficiently using neuromelanin-sensitive MRI. Characterizing neuromelanin signal variations using manual SNc segmentation is an operator-dependent and time-consuming task. Hence, in this cross-sectional, observational, case-control study, we investigated neuromelanin SNc abnormalities in the early PD patients using convolutional neural network-based fully automatic segmentation of SNc. We found a highly significant difference in SNc volume and signal intensity between early PD and healthy volunteers.
|
|||
3255.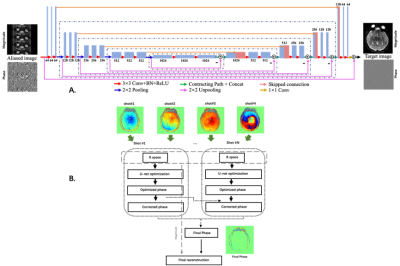 |
Deep learning based high resolution IVIM parameter mapping in lacunar infarction patients
Hui Zhang1, Junqi Xu1, Xutong Kuang2, Shuai Xu2, Xuchen Yu1, Weibo Chen3, Chengyan Wang2, and He Wang1
1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Philips Healthcare, Shanghai, China
This study aimed to propose a reliable deep learning based high resolution intravoxel incoherent motion (IVIM) parameter mapping for the assessment of lacunar infarction patients.
|
|||
3256.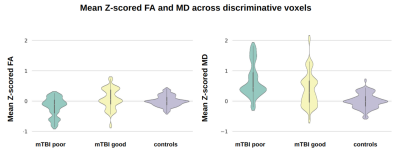 |
Outcome prediction in Mild Traumatic Brain Injury patients using conventional and diffusion MRI via Support Vector Machine: A CENTER-TBI study
Maira Siqueira Pinto1,2, Stefan Winzeck3,4, Marta M. Correia5, Evgenios N. Kornaropoulos4,6, David K. Menon4, Ben Glocker3, Arnold J. den Dekker2, Jan Sijbers2, Pieter-Jan Guns7, Pieter Van Dyck1, and Virginia F. J. Newcombe4
1Radiology, UZA - Antwerp University Hospital, Antwerpen, Belgium, 2imec-Vision Lab, University of Antwerp, Antwerpen, Belgium, 3BioMedIA Group, Department of Computing, Imperial College London, London, United Kingdom, 4Division of Anaesthesia, Department of Medicine, University of Cambridge, Cambridge, United Kingdom, 5MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge, United Kingdom, 6Clinical Sciences, Diagnostic Radiology, Lund University, Lund, Sweden, 7Physiopharmacology, University of Antwerp, Antwerpen, Belgium
Over 40% of patients after a mild traumatic brain injury (mTBI) may have persisting symptoms. This study investigated a Support Vector Machine (SVM) approach for outcome prediction after mTBI from multi-modal MRI. The datasets included 77 mTBI patients from the CENTER-TBI study with acute T2w, SWI, FA and MD scans and outcome scores six month post-injury. Benefits of data harmonization were tested and Z-scoring reduced site-specific biases yielding 67.7% prediction accuracy. Our data-driven approach revealed that predictive signal was retrieved mainly from diffusion maps rather than conventional images, and was located in the superior fronto-occipital fascicle and the corticospinal tract.
|
|||
3257.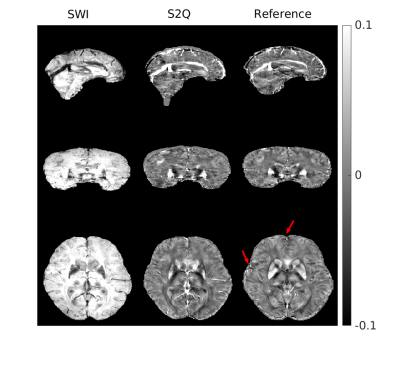 |
Synthesize Quantitative Susceptibility Mapping from Susceptibility Weighting Imaging Using a Cycle Generative Adversarial Network
Zuojun Wang1, Peng Xia1, Henry Ka Fung Mak2, and Peng Cao1
1Diagnostic Radiology, Department of Diagnostic Radiology, HKU, Hong Kong, China, 2Department of Diagnostic Radiology, HKU, Hong Kong, China
Quantitative susceptibility mapping (QSM) obtained from the MRI phase images is valuable in neurological disease diagnoses. Meanwhile, the role of thumb MRI scan probing susceptibility contrast is susceptibility weighting imaging (SWI), which might contain blooming artifacts that would affect the hypointensity appearance. Many conventional methods have been developed for QSM reconstruction, including the deep learning-based approach that is applicable in clinical diagnoses. Here, we apply the cycle generative adversarial network with a perceptual loss to synthesize QSM images from SWI images. The predicted QSM images showed their application in brain microbleed detection.
|
|||
3258.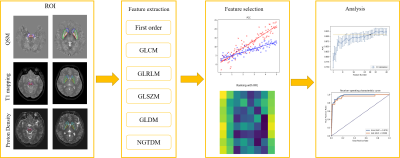 |
Diagnosis of Parkinson’s disease using a radiomics approach based on STrategically Acquired Gradient Echo (STAGE)
Yi Duan1, Yida Wang1, Naying He2, Yan Li2, Zenghui Cheng2, Yu Liu2, Zhijia Jin2, Pei Huang3, Shengdi Chen3, Ewart Mark Haacke2,4, Fuhua Yan2, and Guang Yang1
1East China Normal University, Shanghai Key Laboratory of Magnetic Resonance, Shanghai, China, 2Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 3Department of Neurology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 4Department of Biomedical Engineering, Wayne State University, Detroit, MI, United States
Diagnosing Parkinson’s disease (PD) is still a clinical challenge. Deep grey matter is involved in the pathophysiological changes of PD. We built a radiomics model to distinguish PD from normal controls (NC) based on five brain nuclei in multiple quantitative images derived from STrategically Acquired Gradient Echo (STAGE) imaging. This model combined features from the caudate nucleus, globus pallidus, putamen, red nucleus, and substantia nigra regions in QSM, T1and proton density maps and achieved a test AUC of 0.948. Features from the SN region as seen in the QSM images were found to be the most important ones for classification.
|
|||
3259. |
Prognostic value of MR imaging features derived from automatic segmentation in glioblastoma
Quan Dou1, Xue Feng1, Sohil Patel2, and Craig H. Meyer1
1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Radiology & Medical Imaging, University of Virginia, Charlottesville, VA, United States
Non-invasive MRI-based survival prediction for glioblastoma patients is potentially valuable for informing prognostic and treatment counseling. In this study, we analyzed the relationships between overall survival and several automatic segmentation-based MR imaging features. Simple logistic regression models to classify 1-year survival with clinical factors and selected imaging features were trained and tested. Results showed that combining imaging features with clinical factors improved the survival prediction.
|
|||
3260.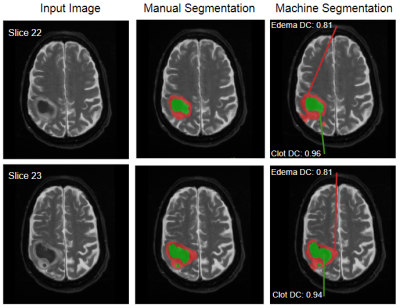 |
Neural Network for Autonomous Segmentation and Volumetric Assessment of Clot and Edema in Intracerebral Hemorrhages
Thomas Lilieholm1, Matt Henningsen2, Azam Ahmed3, Alan McMillan1,4, and Walter F Block1,4,5
1Medical Physics, University of Wisconsin at Madison, Madison, WI, United States, 2Electrical Engineering, University of Wisconsin at Madison, Madison, WI, United States, 3Neurological Surgery, University of Wisconsin at Madison, Madison, WI, United States, 4Radiology, University of Wisconsin at Madison, Madison, WI, United States, 5Biomedical Engineering, University of Wisconsin at Madison, Madison, WI, United States
Previous work has shown that minimally-invasive reduction of hematoma volume in intracerebral hemorrhage to a threshold of 15mL is indicative of improved long term patient outcome. To attain this goal, image-guided minimally-invasive surgical techniques are applied to both lyse clot material and drain from the site of hemorrhage via a porous catheter. We propose a Convolutional Neural Network to identify and autonomously segment clot and peripheral edema in MR images of the brain for volumetric analysis, and image-guidance during evacuation. Quantitative measurements produced in this way can be used for superior clot visualization and direct measurement of remaining clot volume.
|
|||
3261.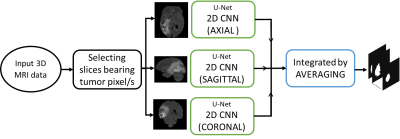 |
A comparative study between multi-view 2D CNN and multi-view 3D anisotropic CNN for brain tumor segmentation
Ritu Lahoti1, Lakshay Agarwal1, Neelam Sinha1, and Vinod Reddy1
1International Institute of Information Technology Bangalore (IIITB), Bengaluru, India Brain tumor segmentation is one of the challenging image segmentation problem. Among the various architectures of CNN used for brain tumor segmentation from MRI images, we compare multi-view 2D CNN and multi-view 3D anisotropic CNN on the popular BraTS dataset. We computed four metrics to measure their performance on 10 test data. The first approach showed a mean sensitivity of 0.816 whereas second approach outperforms with mean sensitivity of 0.9217. Other metrics for both approaches achieved comparable results. Although both approaches consider all orthogonal planes, anisotropic CNN takes into account both global and local features efficiently thereby giving better results. |
|||
3262.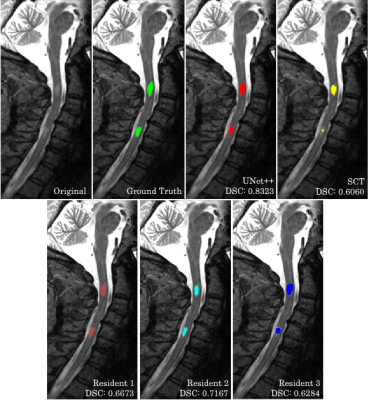 |
Machine Learning Automatic Segmentation of Spinal Cord Lesions in Multiple Sclerosis Patients
Peter Hsu1, Sindhuja Govindarajan1, Nikhil Chettipally1, Lev Bangiyev2, Robert Peyster2, Giuseppe Cruciata2, Patricia Coyle2, Haifang Li2, Hasan Saffiudin1, Ryan Merritt1, Eric Wei1, Almighty Ironnah1, and Kwan Chen1
1Stony Brook University, Stony Brook, NY, United States, 2Stony Brook University Hospital, Stony Brook, NY, United States
Multiple Sclerosis lesions in the spinal cord are associated with more debilitative disease outcomes and have predictive value for prognosis and diagnosis. However, these lesions are difficult to detect from MRI scans and this process is susceptible to inter-rater and intra-rater variability. Machine Learning techniques have the ability to assist in this problem. We propose a Convolutional Neural Network that can perform accurate identification and segmentation of MS lesions in the spinal cord. This method achieves high overlap with the segmentations of attending radiologists and is robust to imaging artifacts, showcasing the potential to be a tool for clinical practice.
|
|||
3263.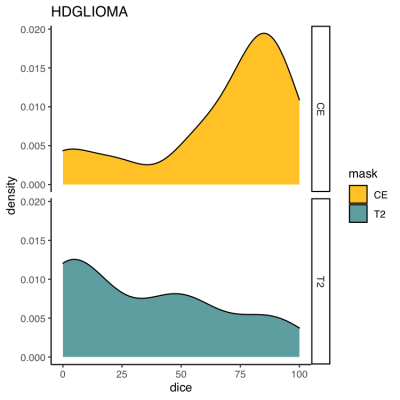 |
What we can learn from adults: Usability of two AI algorithms for Brain and tumor segmentation in a pediatric population.
Maxime DRAI1, GILLES BRUN1, Nadine GIRARD1,2, Benoit TESTUD1,3, and Jan-Patrick STELLMANN1,3
1Neuroradiology, APHM, Marseille, France, 2CRMBM-CEMEREM, Aix-Marseille Université, Marseille, France, 3CNRS, CRMBM-CEMEREM, UMR 7339, Aix-Marseille Université, Marseille, France
AI brain tumor segmentation and brain extraction algorithms are an essential step in image processing, however they are mainly developed in adults. Here, we aimed to explore the usability of these algorithms in a heterogenous pediatric population. In 42 brain pediatric tumor MRI, we compared manual mask with mask generated by the algorithms. Results were excellent for brain extraction, moderate for segmentation of contrast-enhancing tumors, and weak for non-enhancing T2-signal abnormalities. Some improvements are necessary to adapt this algorithm to pediatric brain tumors. However, borrowing strength from adults might be a feasible approach for AI implementation in rare pediatric populations.
|
|||
3264.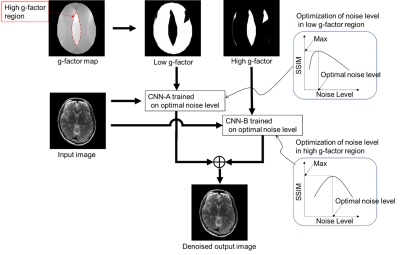 |
Deep-learning-based noise reduction incorporating inhomogeneous spatial distribution of noise in parallel MRI imaging
Atsuro Suzuki1, Chizue Ishihara1, Yukio Kaneko1, Tomoki Amemiya1, Yoshitaka Bito1, and Toru Shirai1
1Healthcare Business Unit, Hitachi, Ltd., Kokubunji-shi, Japan
To reduce inhomogeneous noise caused by parallel imaging, we developed a deep-learning-based noise reduction method that incorporates spatial distribution of noise. For noise distribution we used a g-factor map segmented into high and low g-factor regions. We reduced the noise by using a different optimized network in each region. Finally, a denoised image was generated by combining the two denoised regions. Denoised brain images demonstrated improved signal to noise ratio (SNR) and mean square error (MSE) between denoised and full sampling images throughout the brain regions. Our method was able to reduce the inhomogeneous noise proportional to the noise intensity.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.