Digital Posters
Machine Learning for Image Reconstruction
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 15:00 - 16:00 |
1944.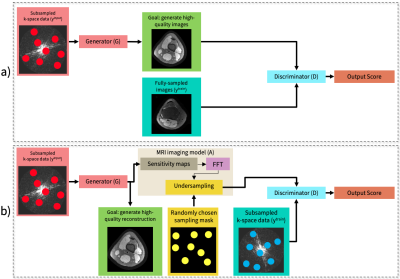 |
Unsupervised Dynamic Image Reconstruction using Deep Generative Adversarial Networks and Total Variation Smoothing
Elizabeth Cole1, Shreyas Vasanawala1, and John Pauly1
1Stanford University, Palo Alto, CA, United States
Deep learning (DL)-based image reconstruction methods have achieved promising results across multiple MRI applications. However, most approaches require large-scale fully-sampled ground truth data for supervised training. Acquiring fully-sampled data is often either difficult or impossible, particularly for dynamic datasets. We present a DL framework for MRI reconstruction which does not use fully-sampled data. We test the proposed method in two scenarios: retrospectively undersampled cine and prospectively undersampled abdominal DCE. Our unsupervised method can produce faster reconstructions which are non-inferior to compressed sensing. Our novel proposed method can enable accelerated imaging and accurate reconstruction in applications where fully-sampled data is unavailable.
|
|||
1945.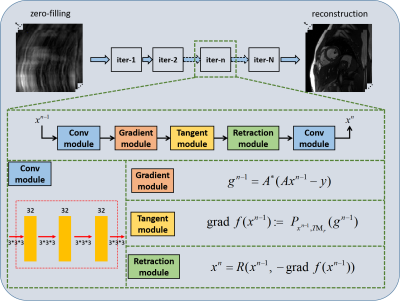 |
Deep Manifold Learning for Dynamic MR Imaging
Ziwen Ke1, Zhuo-Xu Cui1, Jing Cheng1, Leslie Ying2, Xin Liu1, Hairong Zheng1, Yanjie Zhu1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Shenzhen, China, 2University at Buffalo, The State University of New York, Buffalo, NY, United States
Manifold learning has achieved success in cardiac MRI. It models the dynamic images as points on a smooth, low dimensional manifold in high dimensional space. The low dimensional assumption is extracted as a regularizer, but corresponding algorithms are not performed along with the manifold's nonlinear structure. In this paper, we propose a deep manifold learning for dynamic MR imaging. The manifold assumption is no longer taken as the regularization term in the proposed method, but the deep optimization model is directly developed on the nonlinear manifold. The validation on in vivo data shows that our method can achieve improved reconstruction.
|
|||
1946.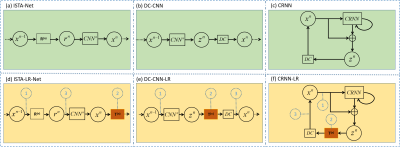 |
A Plug-and-play Low-rank Network Module in Dynamic MR Imaging
Ziwen Ke1, Wenqi Huang1, Jing Cheng1, Leslie Ying2, Xin Liu1, Hairong Zheng1, Yanjie Zhu1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Shenzhen, China, 2University at Buffalo, The State University of New York, Buffalo, NY, United States
Accelerated dynamic MR Imaging is a significant but challenging task. Our previous work demonstrated that deep low-rank priors could achieve improved reconstruction performance by unrolling a sparse and low-rank-based optimization algorithm. However, the optimization algorithm is highly customized, and currently, no deep learning methods exist to apply low-rankness as prior to general inverse problems. In this paper, we propose a plug-and-play low-rank network module in dynamic MR imaging. The low-rank network module can be easily embedded into other deep learning models. The embedding of the LR module can effectively improve the reconstruction results.
|
|||
1947.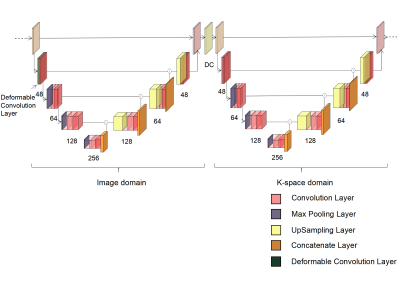 |
Cascaded U-net with Deformable Convolution for Dynamic Magnetic Resonance Imaging
Zhehong Zhang1, Yuze Li2, and Huijun Chen2
1Department of Engineering Physics, Tsinghua University, Beijing, China, 2Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China
The concatenation of several-element U-nets operating in both k-space and image domains is a deep learning network model that has been used for magnetic resonance image (MRI) reconstruction. Here, we present a new method that incorporates deformable 2D convolution kernels into the model. The proposed method leverages motion information of dynamic MRI and thus deformable convolution kernel naturally adapts to image structures. We demonstrate the improved performance of the proposed method using CINE dataset.
|
|||
1948.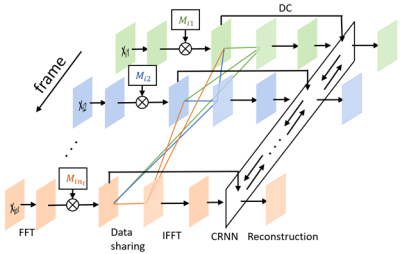 |
Joint deep learning-based optimization of undersampling pattern and reconstruction for dynamic contrast-enhanced MRI
Jiaren Zou1,2 and Yue Cao1,2,3
1Department of Radiation Oncology, University of Michigan, Ann Arbor, MI, United States, 2Department of Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 3Department of Radiology, University of Michigan, Ann Arbor, MI, United States
Joint optimization of deep learning based undersampling pattern and the reconstruction network has shown to improve the reconstruction accuracy for a given acceleration factor in static MRI. Here, we investigate the joint training of a reconstruction network, sampling pattern and data sharing for dynamic contrast-enhanced MRI. By adding a degree of freedom in the temporal direction to the sampling pattern, better reconstruction quality can be achieved. Jointly learned data sharing can further improve the reconstruction accuracy.
|
|||
1949.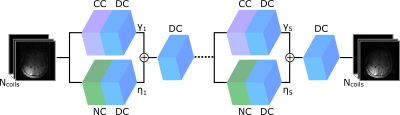 |
A Few-Shot Learning Approach for Accelerated MRI via Fusion of Data-Driven and Subject-Driven Priors
Salman Ul Hassan Dar1,2, Mahmut Yurt1,2, and Tolga Çukur1,2,3
1Department of Electrical and Electronics Engineering, Bilkent University, Ankara, Turkey, 2National Magnetic Resonance Research Center (UMRAM), Bilkent University, Ankara, Turkey, 3Neuroscience Program, Aysel Sabuncu Brain Research Center, Bilkent University, Ankara, Turkey
Deep neural networks (DNNs) have recently found emerging use in accelerated MRI reconstruction. DNNs typically learn data-driven priors from large datasets constituting pairs of undersampled and fully-sampled acquisitions. Acquiring such large datasets, however, might be impractical. To mitigate this limitation, we propose a few-shot learning approach for accelerated MRI that merges subject-driven priors obtained via physical signal models with data-driven priors obtained from a few training samples. Demonstrations on brain MR images indicate that the proposed approach requires just a few samples to outperform traditional parallel imaging and DNN algorithms.
|
|||
1950.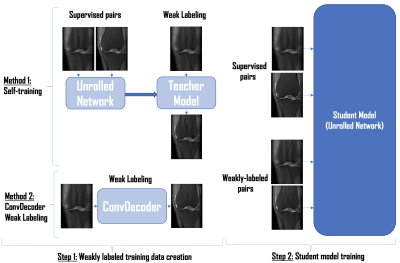 |
Weakly Supervised MR Image Reconstruction using Untrained Neural Networks
Beliz Gunel1, Morteza Mardani1, Akshay Chaudhari2, Shreyas Vasanawala2, and John Pauly1
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Untrained neural networks such as ConvDecoder have emerged as a compelling MR image reconstruction method. Although ConvDecoder does not require any training data, it requires tens of minutes to reconstruct a single MR slice at inference time, making the method impractical for clinical deployment. In this work, we propose using ConvDecoder to construct "weak labels" from undersampled MR scans at training time. Using limited supervised pairs and constructed weakly supervised pairs, we train an unrolled neural network that gives strong reconstruction performance with fast inference time, significantly improving over supervised and self-training baselines in the low data regime.
|
|||
1951.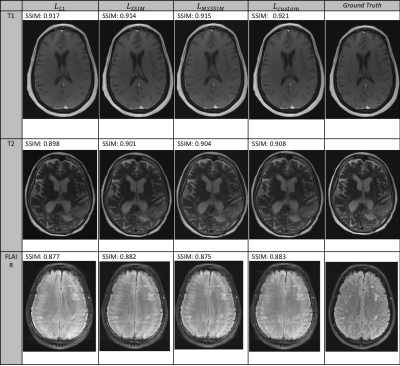 |
A Custom Loss Function for Deep Learning-Based Brain MRI Reconstruction
Abhinav Saksena1, Makarand Parigi1, Nicole Seiberlich2, and Yun Jiang2
1EECS, University of Michigan, Ann Arbor, MI, United States, 2Department of Radiology, University of Michigan, Ann Arbor, MI, United States
The purpose of this work is to test and evaluate a number of candidate loss functions for the reconstruction of diagnostic quality brain MRI images using undersampled k-space data and CNNs. We investigate both per-pixel (L1) and perceptual based (SSIM) loss functions, before developing a custom loss function that incorporates elements of both. We train these loss functions implemented in a UNet architecture on both 4x and 8x undersampled 16-coil MRI data. The custom loss function is shown to produce both the best quantitative results and also sharper and more detailed reconstructions across a number of image contrasts.
|
|||
1952.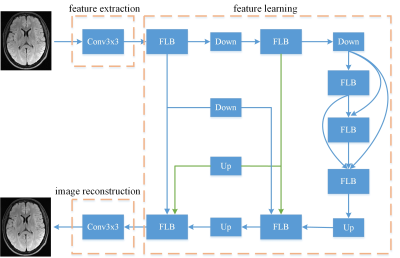 |
A lightweight and efficient convolutional neural network for MR image restoration
Aowen Liu1, Meiling Ji2, Xiaoqian Huang1, Yawei Zhao2, Renkuan Zhai2, Guobin Li2, Dinggang Shen1, and Shu Liao1
1United Imaging Intelligence, Shanghai, China, 2United Imaging Healthcare, Shanghai, China
Many deep learning models for MR image restoration have high computational cost, which raises significant hardware cost and also restricts their usage. To address it, we propose a lightweight network based on the encoder-decoder architecture which integrates image features of different scales and levels to improve the representation capability. A novel loss function is also designed to constrain the model in both image domain and frequency domain. The experimental results show that our model efficiently reduces computational burden while maintaining high performance compared to other conventional models.
|
|||
1953.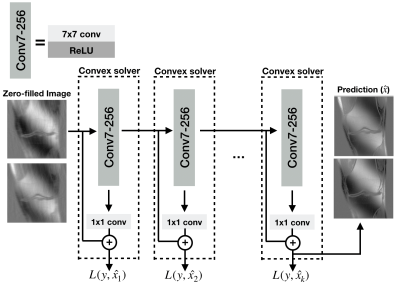 |
Scalable and Interpretable Neural MRI Reconstruction via Layer-Wise Training
Batu Ozturkler1, Arda Sahiner1, Mert Pilanci1, Shreyas Vasanawala2, John Pauly1, and Morteza Mardani1
1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Deep-learning based reconstruction methods have shown great promise for undersampled MR reconstruction. However, their lack of interpretability, and the nonconvex nature impedes their utility as they may converge to undesirable local minima. Moreover, training deep networks in high-dimensional imaging applications such as DCE, and 4D flow requires large amounts of memory that may overload GPUs. Here, we advocate a layer-wise training method amenable to convex optimization, and scalable for training 3D-4D datasets. We compare convex layer-wise training to traditional end-to-end training. The proposed method matches the reconstruction quality of end-to-end training while it is interpretable, convex, and demands less memory.
|
|||
1954.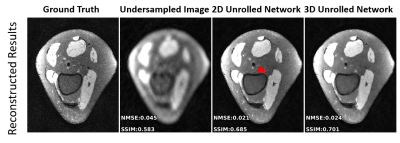 |
Effective Training of 3D Unrolled Neural Networks on Small Databases
Zilin Deng1,2, Burhaneddin Yaman1,2, Chi Zhang1,2, Steen Moeller2, and Mehmet Akçakaya1,2
1University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, Minneapolis, MN, United States
Unrolled neural networks have been shown to improve the reconstruction quality for accelerated MRI. While they have been widely applied in 2D settings, 3D processing may further improve reconstruction quality for volumetric imaging with its ability to capture multi-dimensional interactions. However, implementation of 3D unrolled networks is generally challenging due to GPU-memory limitations and lack of availability of large databases of 3D data. In this work, we tackle both these issues by an augmentation approach that generates smaller sub-volumes from large volumetric datasets. We then compare the 3D unrolled network to its 2D counterpart, showing the improvement from 3D processing.
|
|||
 |
1955.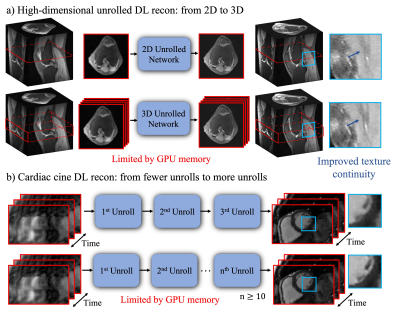 |
Memory-Efficient Learning for High-Dimensional MR Reconstruction
Ke Wang1, Michael Kellman2, Christopher M. Sandino3, Kevin Zhang1, Shreyas S. Vasanawala4, Jonathan I. Tamir5, Stella X. Yu6, and Michael Lustig1
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2Pharmaceutical Chemistry, University of California, San Francisco, Berkeley, CA, United States, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Radiology, Stanford University, Stanford, CA, United States, 5Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States, 6International Computer Science Institute, University of California, Berkeley, Berkeley, CA, United States
High-dimensional Deep learning (DL) reconstructions (e.g. 3D, 2D+time, 3D+time) can exploit multi-dimensional information and achieve improved results over lower dimensional ones. However, the size of the network and its depth for these large-scale reconstructions are currently limited by GPU memory. Here, we use a memory-efficient learning (MEL) framework, which favorably trades off storage with minimal increased computation and enables deeper high-dimensional DL reconstruction on a single GPU. We demonstrate improved image quality with learned high-dimensional reconstruction enabled by MEL for in-vivo 3D MRI and 2D cardiac cine imaging applications. MEL uses much less GPU memory while minimally increasing training time.
|
||
1956.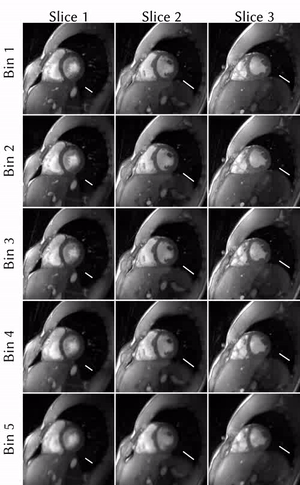 |
Novel insights on SSA-FARY: Amplitude-based respiratory binning in self-gated cardiac MRI
Sebastian Rosenzweig1,2 and Martin Uecker1,2
1Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2Partner Site Göttingen, German Centre for Cardiovascular Research (DZHK), Göttingen, Germany
Cardiac MRI is challenging because of respiratory and cardiac motion. Current clinical approaches try to bypass motion-related issues by ECG-triggering and breath-holds, which comes with several drawbacks. Alternatively, self-gating techniques can be used to determine respiratory and cardiac motion from the acquired raw-data itself. We present novel insights on the quadrature-pair self-gating signals estimated by SSA-FARY: We show that one element of each pair is certain to be in-phase with the motion it represents, as it is the result of a filtering process with a zero-phase filter. This enables the use of less respiratory bins, which decreases the computational demand.
|
|||
1957.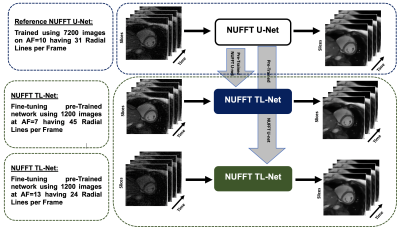 |
Reconstruction of Whole-Heart Cardiac Radial MRI using Neural Network Transfer Learning Approach
Ibtisam Aslam1,2, Fariha Aamir2, Lindsey A CROWE1, Miklos KASSAI1, Hammad Omer2, and Jean-Paul VALLEE1
1Service of Radiology, Geneva University Hospitals and Faculty of Medicine, University of Geneva, Geneva, Switzerland, 2Medical Image Processing Research Group (MIPRG), Deptt. of Electrical & Computer Engineering, COMSATS University Islamabad, Islamabad, Pakistan
In a clinical setting, multiple breath-hold, multi-slice, ECG-gated cine MR (CMR) Cartesian acquisition is a gold standard. Multiple breath-holds in standard CMR acquisition can result in slice-misalignment due to inconsistent breath-hold positions and it forces long exam time. To reduce CMR scan time and to avoid slice-misalignment, under-sampled non-Cartesian (NC) trajectories are useful but lead to artifacts. This paper proposes U-Net based transfer-learning approach with NUFFT (NUFFT TL-Net) to reconstruct artifact-free whole heart, radial CMR images. The preliminary experiments show improved performance of the proposed NUFFT TL-Net both visually and in terms of evaluation parameters than contemporary NUFFT U-Net.
|
|||
1958.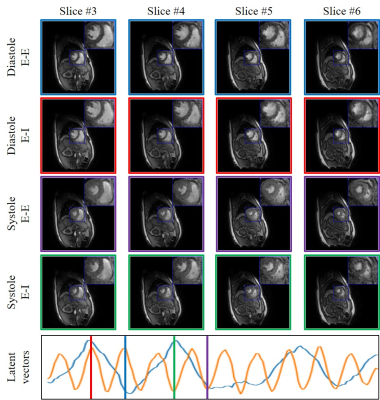 |
Alignment & joint recovery of multi-slice cine MRI data using deep generative manifold model
Qing Zou1, Abdul Haseeb Ahmed1, Prashant Nagpal1, Rolf Schulte2, and Mathews Jacob1
1University of Iowa, Iowa City, IA, United States, 2GE Global Research, Munich, Germany The main focus of this work is to introduce an unsupervised deep generative manifold model for the alignment and joint recovery of the slices in free-breathing and ungated cardiac cine MRI. The main highlights are (1) the ability to align multi-slice data and capitalize on the redundancy between the slices. (2) The ability to estimate the gating information directly from the k-t space data. (3) The unsupervised learning strategy that eliminates the need for extensive training data. The joint recovery facilitates the acquisition of data from the whole heart in around 2 minutes of acquisition time. |
|||
1959.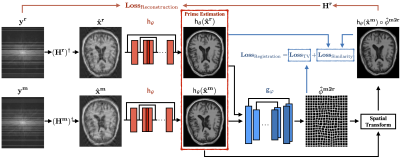 |
Deep image reconstruction for MRI using unregistered measurement pairs without ground truth
Weijie Gan1, Yu Sun1, Cihat Eldeniz2, Jiaming Liu3, Hongyu An2, and Ulugbek S. Kamilov1,3
1Department of Computer Science and Engineering, Washington University in St. Louis, St. Louis, MO, United States, 2Mallinckrodt Institute of Radiology, Washington University in St. Louis, St. Louis, MO, United States, 3Department of Electrical and Systems Engineering, Washington University in St. Louis, St. Louis, MO, United States
One of the key limitations in conventional deep learning-based MR image reconstruction is the need for registered pairs of training images, where fullysampled ground truth images are required as the target. We address this limitation by proposing a novel registration-augmented image reconstruction method that trains a CNN by directly mapping pairs of unregistered and undersampled MR measurements. The proposed method is validated on a single-coil MRI data set by training a model directly on pairs of undersampled measurements from images that have undergone nonrigid deformations.
|
|||
1960.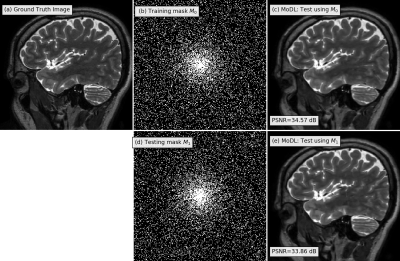 |
Adaptive deep image reconstruction using G-SURE
Hemant Kumar Aggarwal1 and Mathews Jacob1
1University of Iowa, Iowa City, IA, United States Deep learning image reconstruction algorithms often suffer from model mismatches when the acquisition scheme differs significantly from the forward model used during training. We introduce a Generalized Stein's Unbiased Risk Estimate (GSURE) loss metric to adapt or fine-tune the network to the measured k-space data, thus minimizing the impact of model misfit. Unlike current methods that rely on the mean square error in k-space, the proposed metric accounts for noise in the measurements. This makes the approach less vulnerable to overfitting, thus offering improved reconstruction quality compared to schemes that rely on mean-square error. |
|||
1961.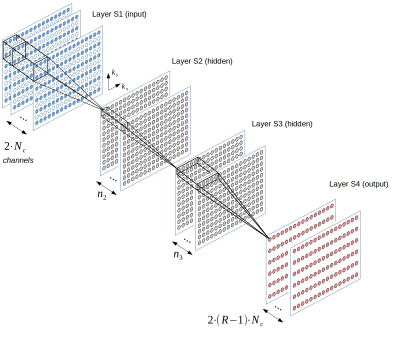 |
Influence of training data on RAKI reconstruction quality in standard 2D imaging
Peter Dawood1,2, Martin Blaimer3, Peter M. Jakob1, and Johannes Oberberger2
1Department of Experimental Physics 5, University of Würzburg, Würzburg, Germany, 2Department of Internal Medicine I, University Hospital Würzburg, Würzburg, Germany, 3Magnetic Resonance and X-Ray Imaging Department, Development Center X-ray Technology EZRT, Fraunhofer Institute for Integrated Circuits IIS, Würzburg, Germany
The parallel imaging method GRAPPA has been generalized within the Machine Learning framework by introducing the deep-learning method RAKI, in which Convolutional Neural Networks are used for non-linear k-space interpolation. RAKI is a database-free approach that uses scan-specific calibration data. Here, we study the influence of the calibration data on the image quality of 2D imaging sequences. The results indicate that RAKI yields superior signal-to-noise ratio but introduces blurring and loss of detail for typical calibration data amounts at high accelerations. Furthermore, the contrast information in the calibration data must be similar to that of the accelerated scans.
|
|||
1962. |
Non-uniform Fast Fourier Transform via Deep Learning
Yuze Li1, Zhangxuan Hu2, Haikun Qi3, Guangqi Li1, Dongyue Si1, Haiyan Ding1, Hua Guo1, and Huijun Chen1
1Center for Biomedical Imaging Research, Medical School, Tsinghua University, Beijing, China, 2MR Research China, GE Healthcare, Beijing, China, 3King’s College London, London, United Kingdom
In this study, a deep learning-based MR reconstruction framework called DLNUFFT (Deep Learning-based Non-Uniform Fast Fourier Transform) was proposed, which can restore the under-sampled non-uniform k-space to fully sampled Cartesian k-space without NUFFT gridding. Novel network blocks with fully learnable parameters were built to replace the hand-crafted convolution kernel and the density compensation in NUFFT. Simulations and in-vivo results showed DLNUFFT can achieve higher performance than conventional NUFFT, compressed sensing and state-of-the-art deep learning methods in terms of PSNR and SSIM.
|
|||
1963.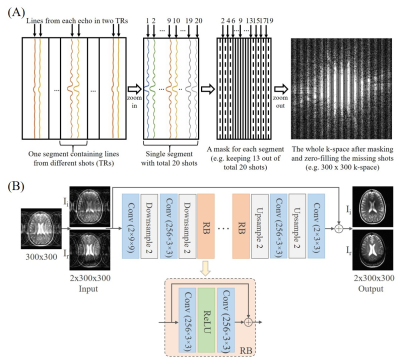 |
Deep Learning Image Reconstruction from Incomplete Fast Spin Echo MR Data
Linfang Xiao1,2, Yilong Liu1,2, Yujiao Zhao1,2, Zheyuan Yi1,2,3, Vick Lau1,2, Alex T.L. Leong1,2, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
Fast spin echo (FSE) is the most commonly used multi-shot sequence in clinical MRI. In this study, we propose to acquire single-channel FSE data with incomplete number of shots (TRs), and reconstruct such periodically undersampled k-space data using a deep learning approach. The results demonstrate that the proposed method can effectively remove the aliasing artifacts and recover the high frequency information without noise amplification, enabling a FSE acceleration that can be readily implemented in practice.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.