Digital Posters
Machine Learning for Quantitative Imaging
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 17:00 - 18:00 |
2162.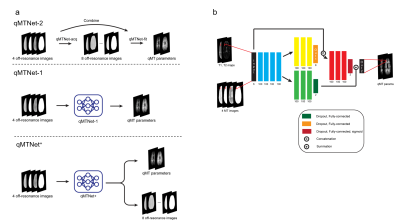 |
qMTNet+: artificial neural network with residual connection for accelerated quantitative magnetization transfer imaging
Huan Minh Luu1, Dong-Hyun Kim1, Seung-Hong Choi2, and Sung-Hong Park1
1Magnetic Resonance Imaging Laboratory, Department of Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Seoul National University Hospital, Seoul, Korea, Republic of
Quantitative magnetization transfer (qMT) imaging provides quantitative measures of magnetization transfer properties, but the method itself suffers from long acquisition and processing time. Previous research has looked into the application of deep learning to accelerate qMT imaging. Specifically, a network called qMTNet was proposed to accelerate both data acquisition and fitting. In this study, we propose qMTNet+, an improved version of qMTNet, that accomplishes both acceleration tasks as well as generation of missing data with a single residual network. Results showed that qMTNet+ improves the quality of generated MT images and fitted qMT parameters compared to qMTNet.
|
|||
2163.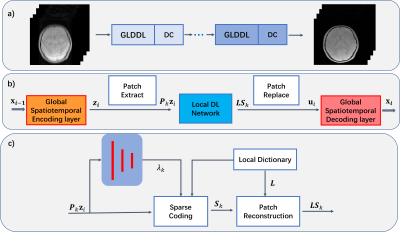 |
Global and Local Deep Dictionary Learning Network for Accelerating the Quantification of Myelin Water Content
Quan Chen1, Huajun She1, Zhijun Wang1, and Yiping P. Du1
1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Acceleration of the myelin water fraction (MWF) mapping at R=6 using a Global and Local Deep Dictionary Learning Network (GLDDL) is demonstrated in this study. The global and the local spatiotemporal correlations in the relaxations are learned simultaneously. The global temporal encode-decoder layers are utilized to reduce the computational complexity of the local DL network and improve the denoising performance. The deep DL network utilizes the merits of traditional DL and deep learning to improve the reconstruction. The high-quality MWF maps obtained from the GLDDL network has demonstrated the feasibility to accelerate the whole brain MWF mapping in 1 minute.
|
|||
2164.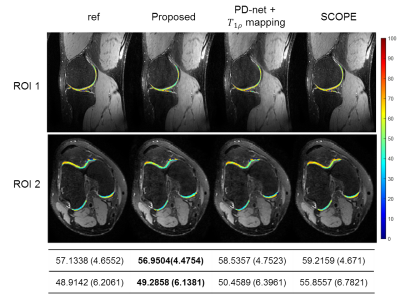 |
Rapid MR Parametric Mapping using Deep Learning
Jing Cheng1, Yuanyuan Liu1, Xin Liu1, Hairong Zheng1, Yanjie Zhu1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Existing deep learning-based methods for rapid MR parametric mapping often use the reference parametric maps fitted from fully sampled images to train the networks. Nevertheless, the fitted parametric map is sensitive to the noise and the fitting algorithms. In this work, we proposed to incorporate the quantitative physical model into the deep learning framework to simultaneously reconstruct the parameter-weighted images and generate the parametric map without the reference parametric maps. Experimental results on the quantitative MR T1ρ mapping show the promising performance of the proposed framework.
|
|||
2165.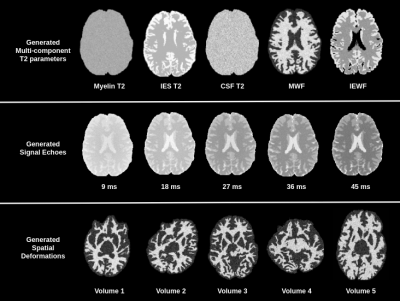 |
Synthesizing large scale datasets for training deep neural networks in quantitative mapping of myelin water fraction
Serge Vasylechko1,2, Simon K. Warfield1,2, Sila Kurugol1,2, and Onur Afacan1,2
1Boston Children's Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
Deep learning methods have the potential to improve quantitative MRI methods. However, performance of deep learning methods is highly sensitive to the amount of available training data. In this work, we propose generating a substantial amount of 3D synthetic data, and demonstrate its application to myelin water fraction mapping. A parameter sampling model is designed within a naturally occurring range of multi-component T2 distributions to generate a large set of varying synthetic signals. This model is combined with a spatially varying sampling model that generates a multitude of spatial deformations and signal perturbations.
|
|||
2166.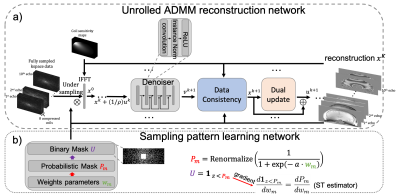 |
Deep unrolled network with optimal sampling pattern to accelerate multi-echo GRE acquisition for quantitative susceptibility mapping
Jinwei Zhang1, Hang Zhang1, Pascal Spincemaille2, Mert Sabuncu3, Thanh Nguyen2, Ilhami Kovanlikaya2, and Yi Wang2
1Cornell University, New York, NY, United States, 2Weill Cornell Medical College, New York, NY, United States, 3Cornell University, Ithaca, NY, United States
To accelerate the acquisition time of quantitative susceptibility mapping (QSM) using a 3D multi-echo gradient echo (mGRE) sequence, an unrolled multi-channel deep ADMM reconstruction network with a LOUPE-ST based 2D variable density sampling pattern optimization module is trained to optimize both the k-space under-sampling pattern and the reconstruction. Prospectively under-sampled k-space data are acquired using a modified mGRE sequence and reconstructed by the trained unrolled network. Prospective study shows the learned sampling pattern achieves better image quality in QSM compared to a manually designed pattern.
|
|||
2167. |
Automated quantitative evaluation of deep learning model for reduced gadolinium dose in contrast-enhanced brain MRI
Srivathsa Pasumarthi1, Jon Tamir2, Enhao Gong2, Greg Zaharchuk2, and Tao Zhang2
1Subtle Medical Inc, Menlo Park, CA, United States, 2Subtle Medical Inc., Menlo Park, CA, United States
Deep learning (DL) has recently become a promising technique to address the safety concerns for Gadolinium-based Contrast Agents (GBCAs) in MRI. Studies have shown that high quality contrast-enhanced images can be generated by DL with only a small fraction of the standard dose, and in some cases no dose at all. To build upon existing research that has heavily focused on qualitative evaluation by radiologists, this work proposes an automated quantitative evaluation scheme for the GBCA dose reduction using DL.
|
|||
2168.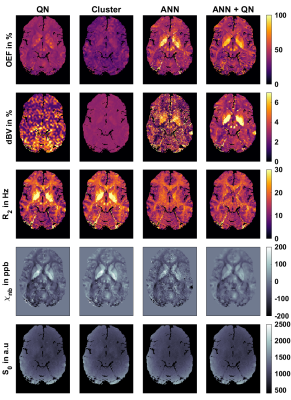 |
Using an ANN to estimate Initial Values for Mapping of the Oxygen Extraction Fraction with combined QSM and qBOLD
Patrick Kinz1, Sebastian Thomas1, and Lothar R. Schad1
1Computer Assisted Clinical Medicine, Heidelberg University, Medical Faculty Mannheim, Mannheim, Germany
MRI-based mapping of oxygen extraction fraction with QSM and qBOLD is a non-invasive diagnostic tool with many possible applications. But current reconstruction methods based on quasi-Newton (QN) methods are very dependent on accurate parameter initialization. We compare the standard QN method with a clustering approach, and with a new initialization method in form of an ANN, which we previously developed to replace QN. This leads to reconstructed images with much less noise, compared to the direct output of the ANN and keeps the effect of reduced intersubject variability.
|
|||
2169.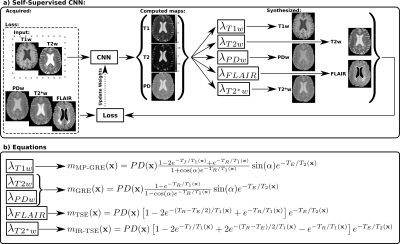 |
A self-supervised deep learning approach to synthesize weighted images and T1, T2, and PD parametric maps based on MR physics priors
Elisa Moya-Sáez1,2, Rodrigo de Luis-García1, and Carlos Alberola-López1
1University of Valladolid, Valladolid, Spain, 2Fundación Científica AECC, Valladolid, Spain
Synthetic MRI is gaining popularity due to its ability to generate realistic MR images. However, T1, T2, and PD maps are rarely used despite they provide the information needed to synthesize any image modality by applying the appropriate theoretical equations derived from MR physics or through involved simulation procedures. In this work we propose an extension of an state-of-the-art standard CNN to a self-supervised CNN by including MR physics priors to tackle confounding factors not considered in the equations, while bypassing the need of costly simulations. Our approach yields both realistic maps and weighted images from real data.
|
|||
2170.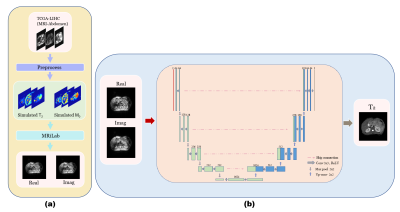 |
Free-breathing Abdomen T2 mapping via Single-shot Multiple Overlapping-echo Acquisition and Deep Neural Network Reconstruction
Xi Lin1, Qinqin Yang1, Jianfeng Bao2, Shuhui Cai1, Zhong Chen1, Congbo Cai1, and Jingliang Cheng2
1Department of Electronic Science, Xiamen University, Xiamen, China, 2Department of Magnetic Resonance Imaging, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, China
Quantitative T2 mapping of abdomen has the potential for many clinical scenarios, such as for distinguishing cirrhosis and characterizing renal lesions. In this study, we applied overlapping echo acquisition together with deep learning-based reconstruction to achieving T2 mapping of abdomen in single shot for the first time. The resulting abdominal T2 values are in good agreement with the reported ones. This method makes high temporal resolution quantitative abdominal imaging possible, and has strong motion robustness.
|
|||
2171.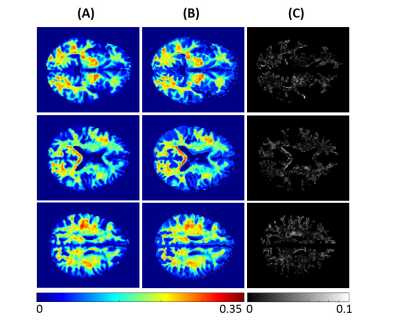 |
Myelin water fraction determination from relaxation times and proton density through deep learning neural network
Nikkita Khattar1, Zhaoyuan Gong1, Matthew Kiely1, Curtis Triebswetter1, Maryam H. Alsameen1, and Mustapha Bouhrara1
1Laboratory of Clinical Investigation, National Institute on Aging, Baltimore, MD, United States
MRI mapping of myelin water fraction (MWF), a surrogate of myelin content, has provided important insights into brain maturation and neurodegeneration, with promising potential use to quantify disease progression or therapeutic effect. Besides the complex modeling, MWF imaging, using either conventional or advanced methods such as the BMC-mcDESPOT approach, requires prolonged acquisition times, hampering their integration in clinical investigations. In this proof-of-concept work, we propose an artificial neural network model to derive MWF maps from conventional relaxation times and proton density maps. This work opens a way to further developments for practical and fast MWF imaging.
|
|||
2172.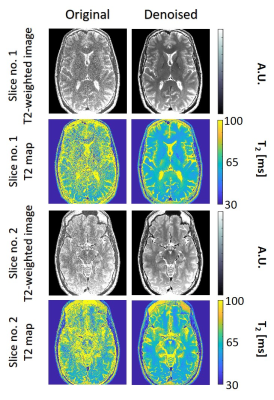 |
In-Vivo evaluation of high resolution T2 mapping using Bloch simulations and MP-PCA image denoising
Neta Stern1, Dvir Radunsky1, Tamar Blumenfeld-Katzir1, Yigal Chechik2,3, Chen Solomon1, and Noam Ben-Eliezer1,4,5
1Department of Biomedical Engineering, Tel Aviv University, Tel Aviv, Israel, 2Department of Orthopedics, Shamir Medical Center, Zerifin, Israel, 3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 4Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 5Center for Advanced Imaging Innovation and Research (CAI2R), New-York University, Langone Medical Center, NY, United States
This study evaluates the performance of Marchenko-Pastur Principle Component Analysis (MP-PCA) denoising algorithm for improving T2 mapping precision using the EMC algorithm. Denoising efficiency was successfully validated on in vivo brain and knee scans, showing an increase in T2 maps’ precision while preserving anatomical features. This enables precise mapping of T2 values at high-resolutions. The proposed method can benefit a wide range of clinical applications, and carries the potential to enhance the detection of abnormalities on both T2 weighted images and qualitative T2 maps.
|
|||
2173.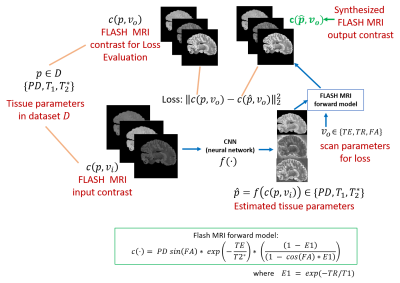 |
Rapid learning of tissue parameter maps through random FLASH contrast synthesis
Divya Varadarajan1,2, Katie Bouman3, Bruce Fischl*1,2,4, and Adrian Dalca*1,5
1Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Computing and Mathematical Sciences, California Institute of Technology, Pasadena, CA, United States, 4Massachusetts General Hospital, Boston, MA, United States, 5Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States
Estimating tissue properties and synthesizing contrasts can ease the need for long acquisitions and have many clinical applications. In this work we propose an unsupervised deep-learning strategy that employs the FLASH MRI model. The method jointly estimates the T1, T2* and PD tissue parameter maps with the goal to synthesize physically plausible FLASH signals. Our approach is additionally trained for random acquisition parameters and generalizes across different acquisition protocols and provides improved performance over fixed acquisition based training methods. We also demonstrate the robustness of our approach by performing these estimation with as low as three input contrast images.
|
|||
2174.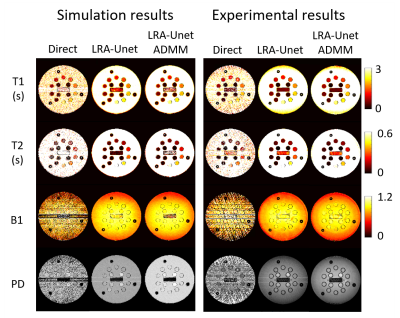 |
Accurate quantitative parameter maps reconstruction method for tsDESPOT using Low Rank approximated Unet ADMM
Yuuzo Kamiguchi1, Sadanori Tomiha2, and Masao Yui3
1Advanced Technology Reserch Dept. Reserch and Development Center, Canon Medical Systems Corporation, Kawasaki, Japan, 2Advanced Technology Reserch Dept. Reserch and Development Center, Canon Medical Systems Corporation, Otawara, Japan, 3Reserch and Development Center, Canon Medical Systems Corporation, Otawara, Japan
We examined the quantitative multi parameter mapping method so called transient state DESPOT (tsDESPOT) which based on conventional DESPOT sequence. From the acquired data, low rank approximated (LRA) images which include transient state information were reconstructed, then T1, T2, B1 and PD maps were estimated by dense neural network. In this study, we proposed fast estimation method of accurate full sampled LRA images using approximate ADMM (alternating direction method of multipliers) which optimize Unet estimation and data consistency. Compared to simple Unet estimation method, the method improved quantitative accuracy of maps and removed artifact that couldn’t be removed.
|
|||
2175.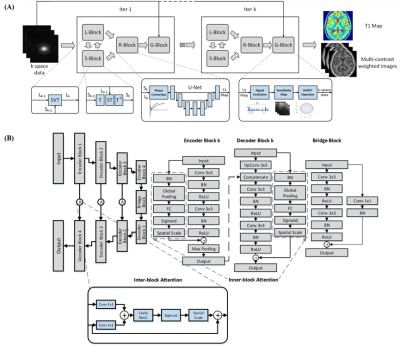 |
Deep Learning Enhanced T1 Mapping and Reconstruction Framework with Spatial-temporal and Physical Constraint
Yuze Li1, Huijun Chen1, Haikun Qi2, Zhangxuan Hu3, Zhensen Chen1, Runyu Yang1, Huiyu Qiao1, Jie Sun4, Tao Wang5, Xihai Zhao1, Hua Guo1, and Huijun Chen1
1Center for Biomedical Imaging Research, Medical School, Tsinghua University, Beijing, China, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3GE Healthcare, Beijing, China, 4Vascular Imaging Lab and BioMolecular Imaging Center, Department of Radiology, University of Washington, Seattle, Seattle, WA, United States, 5Department of Neurology, Peking University Third Hospital, Beijing, China
A Deep learning enhAnced T1 parameter mappIng and recoNstruction framework using spatial-Temporal and phYsical constraint (DAINTY) was proposed. DAINTY explicitly imposed low rank and sparsity constraints on the multi-frame T1 weighted images to exploit the spatial-temporal correlation. A deep neural network was used to efficiently perform T1 mapping as well as denoise and reduce under-sampling artifacts. More importantly, smooth and accurate T1 maps generated from the neural network were transformed to T1 weighted images using the physical model, which the transformed T1 weighted images were also refined. Combining refined images and intermediate reconstructed images, the image quality was greatly improved.
|
|||
2176.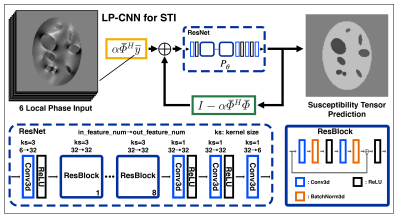 |
Learned Proximal Convolutional Neural Network for Susceptibility Tensor Imaging
Kuo-Wei Lai1,2, Jeremias Sulam1, Manisha Aggarwal3, Peter van Zijl2,3, and Xu Li2,3
1Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Radiology and Radiological Sciences, Johns Hopkins University, Baltimore, MD, United States
We extended a Learned Proximal Convolutional-Neural-Network (LP-CNN) model used in scalar-based Quantitative Susceptibility Mapping (QSM) to tensor-based Susceptibility Tensor Imaging (STI). To improve the accuracy in reconstructed susceptibility anisotropy and tensor eigenvectors, we devised a decomposition loss function to balance training errors in isotropic and anisotropic components. Results using a synthetic dataset demonstrated that, compared to the conventional iterative approach, LP-CNN-STI provides better estimates of susceptibility tensor and smaller errors in anisotropy and eigenvectors. This deep learning-based STI method naturally incorporates the STI physical model, and is a first step toward development of learning-based STI potentially with less acquisition orientations.
|
|||
2177.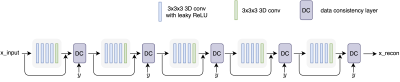 |
Accelerating 3D MULTIPLEX MRI Reconstruction with Deep Learning
Eric Z. Chen1, Yongquan Ye2, Xiao Chen1, Jingyuan Lyu2, Zhongqi Zhang3, Yichen Hu2, Terrence Chen1, Jian Xu2, and Shanhui Sun1
1United Imaging Intelligence, Cambridge, MA, United States, 2UIH America, Inc., Houston, TX, United States, 3United Imaging Healthcare, Shanghai, China
We propose a deep learning framework for undersampled 3D MRI data reconstruction and apply it to a recently developed multi-flip-angle (FA) and multi-echo GRE method (named MULTIPLEX) that can simultaneously acquire multiple parametric images with just one single MRI scan. The proposed deep learning method shows good performance in image quality and reconstruction time.
|
|||
2178.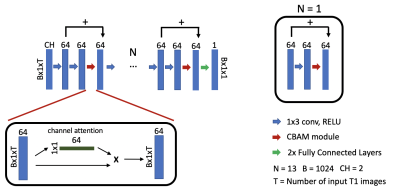 |
Accelerated cardiac T1 mapping using attention-gated neural networks
Johnathan Le1,2, Jason Mendes2, Mark Ibrahim3, Brent Wilson3, Edward DiBella1,2, and Ganesh Adluru1,2
1Department of Biomedical Engineering, University of Utah, Salt Lake City, UT, United States, 2Utah Center for Advanced Imaging Research, University of Utah, Salt Lake City, UT, United States, 3Department of Cardiology, University of Utah, Salt Lake City, UT, United States
Cardiac T1 mapping has been shown to be a promising method for assessing different cardiomyopathies. Recently, native T1 mapping has been used to identify ischemic regions in coronary artery disease without the use of gadolinium-based contrast agents. Most cardiac T1 mapping methods require long breath holds during the acquisition sequence which can be difficult for patients particularly during exercise or pharmacologically induced stress. Here we proposed using attention-gated neural networks to reduce the acquisition time of native and post-contrast cardiac T1 mapping sequences without significant loss of quality.
|
|||
2179.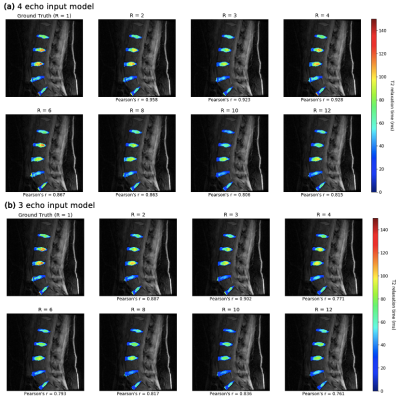 |
8X Accelerated Intervertebral Disc Compositional Evaluation with Recurrent Encoder-Decoder Deep Learning Network
Aniket Tolpadi1,2, Francesco Caliva1, Misung Han1, Valentina Pedoia1, and Sharmila Majumdar1
1Radiology and Biomedical Imaging, UCSF, San Francisco, CA, United States, 2Bioengineering, University of California, Berkeley, Berkeley, CA, United States
Degeneration of intervertebral discs (IVDs) is correlated with low back pain, but conventional MRI fails to capture early signs of degeneration. Quantitative MRI (qMRI) is sensitive to early degenerative biochemical changes but suffers from long acquisition times. We present a recurrent encoder-decoder architecture that predicts fully sampled IVD T2 maps from spatially and temporally undersampled qMRI echos. The network allows for up to eightfold reduction in acquisition time while exhibiting strong correlation to ground truth maps, maintaining fidelity to T2 values, and retaining textures. With further development, this network can make qMRI a more regular part of lumbar spine imaging.
|
|||
2180.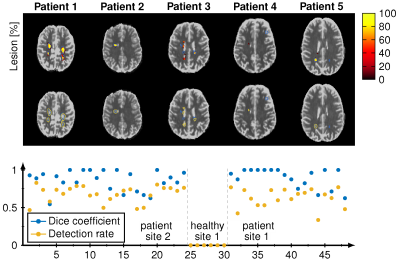 |
Deep Learning Reconstruction of MR Fingerprinting for simultaneous T1, T2* mapping and generation of WM, GM and WM lesion probability maps
Ingo Hermann1,2,3, Alena-Kathrin Golla1,3, Eloy Martinez-Heras4, Ralf Schmidt1, Elisabeth Solana4, Sara Llufriu4, Achim Gass5, Lothar Schad1, Sebastian Weingärtner2, and Frank Zöllner1,3
1Computer Assisted Clinical Medicine, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany, 2Magnetic Resonance Systems Lab, Department of Imaging Physics, Delft University of Technology, Delft, Netherlands, 3Mannheim Institute for Intelligent Systems in Medicine, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany, 4Center of Neuroimmunology, Laboratory of Advanced Imaging in Neuroimmunological Diseases, Hospital Clinic Barcelona, Universitat de Barcelona, Barcelona, Spain, 5Department of Neurology, Medical Faculty Mannheim, University Heidelberg, Mannheim, Germany
A single deep learning regression network was trained for unified reconstruction, distortion correction and denoising of T1, T2*, WM, GM and lesion probability maps from MRF-EPI acquisition. The network was trained with binary lesion masks and the WM, GM probability maps generated with SPM. The training T1 and T2* maps were reconstructed using dictionary matching. The relative deviation was 7.6% for the 5 output mask in the whole brain between the proposed deep learning network and the conventional processing. Dice coefficients were 0.85 for WM and GM and 0.67 for the lesions with a lesion detection rate of 0.83.
|
|||
 |
2181.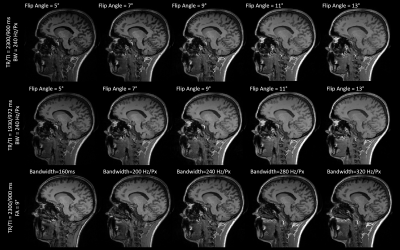 |
The sensitivity of classical and deep image similarity metrics to MR acquisition parameters
Veronica Ravano1,2,3, Gian Franco Piredda1,2,3, Tom Hilbert1,2,3, Bénédicte Maréchal1,2,3, Reto Meuli2, Jean-Philippe Thiran2,3, Tobias Kober1,2,3, and Jonas Richiardi2
1Advanced Clinical Imaging Technology, Siemens Healthineers, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Post-acquisition data harmonization promises to unlock multi-site data for deep learning applications. In turn, this rests on measuring image similarity. Here, we investigate the sensitivity of several similarity measures to fourteen acquisition protocols of a 3D T1-weighted (MPRAGE) contrast. Standard similarity metrics, a deep perceptual loss and a segmentation loss are extracted between image pairs and compared. The perceptual loss is highly correlated with L1 distance and outperforms other metrics in detecting acquisition parameter changes. The segmentation loss, however, is poorly correlated with other metrics, suggesting that these image similarity metrics alone aren't sufficient to harmonize data for clinical applications.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.