Oral
Constrained & Model-Based Reconstructions
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 14:00 - 16:00 | Moderators: Merry Mani & Haifeng Wang |
 |
0063.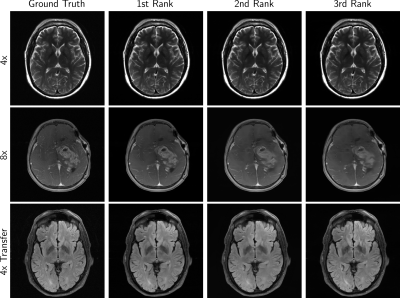 |
Results of the 2020 fastMRI Brain Reconstruction Challenge
Bruno Riemenschneider1, Matthew Muckley2, Alireza Radmanesh1, Sunwoo Kim3, Geunu Jeong3, Jingyu Ko3, Yohan Jun4, Hyungseob Shin4, Dosik Hwang4, Mahmoud Mostapha5, Simon Arberet5, Dominik Nickel6, Zaccharie Ramzi7,8, Philippe
Ciuciu7, Jean-Luc Starck7, Jonas Teuwen9, Dimitrios Karkalousos10, Chaoping Zhang10, Anuroop Sriram11, Zhengnan Huang1, Nafissa Yakubova2, Yvonne W. Lui1, and Florian Knoll1
1NYU School of Medicine, New York, NY, United States, 2Facebook AI Research, New York, NY, United States, 3AIRS Medical, Seoul, Korea, Republic of, 4Yonsei University, Seoul, Korea, Republic of, 5Siemens Healthineers, Princeton, NJ, United States, 6Siemens Healthcare GmbH, Erlangen, Germany, 7CEA (NeuroSpin) & Inria Saclay (Parietal), Université Paris-Saclay, Gif-sur-Yvette, France, 8Département d’Astrophysique, CEA-Saclay, Gif-sur-Yvette, France, 9Radboud University Medical Center, Nijmegen, Netherlands, 10Amsterdam UMC, Amsterdam, Netherlands, 11Facebook AI Research, Menlo Park, CA, United States
The next round of the fastMRI reconstruction challenge took place, this time using anatomical brain data. Submissions were ranked by SSIM and resulting finalists again by 6 radiologists. We observed the cases with clear SSIM separation achieving the highest radiologists’ rankings, in particular the winning reconstructions. Most 4x track reconstructions exhibit desirable image quality, with some exceptions that show anatomy-like hallucinations. Radiologist sentiment decreased for the 8x and Transfer tracks, indicating that these may require further investigation.
|
|
 |
0064.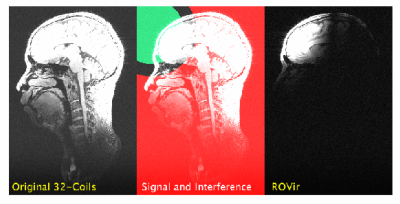 |
Region-Optimized Virtual (ROVir) Coils: Application of Sensor-Domain Beamforming for Localizing and/or Suppressing Spatial Regions
Daeun Kim1, Stephen F. Cauley2, Krishna S. Nayak1, Richard M. Leahy1, and Justin P. Haldar1
1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Radiology, A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States
MRI acquisitions often incidentally excite spatial regions that are not interesting for the application. This unnecessary magnetization can lead to artifacts and/or prolonged acquisitions. We propose a novel virtual-coil approach, called region-optimized virtual (ROVir) coils, that can localize signal from an ROI and/or suppress signal from unwanted spatial regions while also providing coil compression. This is achieved using optimal beamforming principles (without requiring modification of pulse sequences or imaging hardware), and can be applied directly to k-space data, which enables simplified image reconstruction. We illustrate ROVir with reduced-FOV imaging, demonstrating capabilities to suppress aliasing artifacts from outside the nominal FOV.
|
|
 |
0065.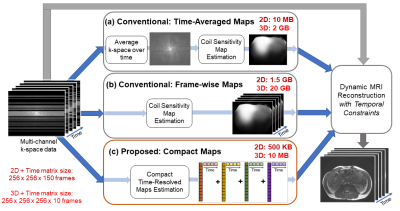 |
Compact Maps: A Low-Dimensional Approach for High-Dimensional Time-Resolved Coil Sensitivity Map Estimation
Shreya Ramachandran1, Frank Ong2, and Michael Lustig1
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2Electrical Engineering, Stanford University, Stanford, CA, United States
Dynamic MRI reconstruction techniques often use static coil sensitivity maps, but physical sensitivities can change substantially with respiratory and other subject motion. However, time-resolved sensitivity maps occupy a very large amount of memory, and hence, directly employing these maps is often impractical, especially on memory-limited GPUs. Here, we introduce a technique that solves for a compact representation of time-resolved sensitivity maps by leveraging a temporal basis for sensitivity kernels. Our proposed Compact Maps are significantly cheaper (by ~1000x) to store in memory than conventional time-resolved maps and result in lower calibration error and reconstruction error than do time-averaged maps.
|
|
 |
0066.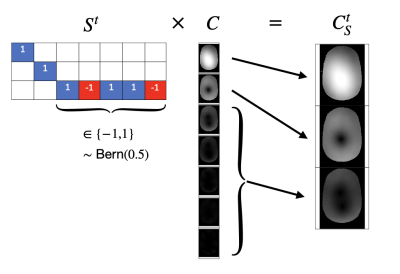 |
Coil Sketching for fast and memory-efficient iterative reconstruction
Julio A. Oscanoa1,2, Frank Ong3, Zhitao Li2,3, Christopher M. Sandino3, Daniel B. Ennis2,4, Mert Pilanci3, and Shreyas S. Vasanawala2
1Department of Bioengineering, Stanford University, Stanford, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 4Cardiovascular Institute, Stanford, CA, United States
Parallel imaging and compressed sensing reconstruction of large datasets has a high computational cost, especially for 3D non-Cartesian acquisitions. This work is motivated by the success of iterative Hessian sketching methods in machine learning. Herein, we develop Coil Sketching to lower computational burden by effectively reducing the number of coils actively used during iterative reconstruction. Tested with 2D radial and 3D cones acquisitions, our method yields considerably faster reconstructions (around 2x) with virtually no penalty on reconstruction accuracy.
|
|
 |
0067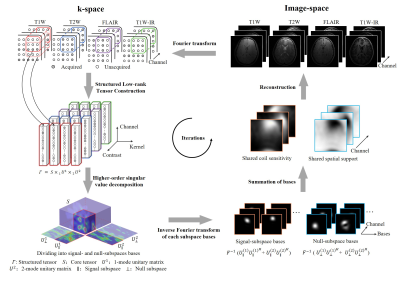 |
Fast Calibrationless Image-space Reconstruction by Structured Low-rank Tensor Estimation of Coil Sensitivity and Spatial Support Video Permission Withheld
Zheyuan Yi1,2,3, Yujiao Zhao1,2, Yilong Liu1,2, Yang Gao1,2, Mengye Lyu4, Fei Chen3, and Ed X Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China, 4College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, China
In conventional parallel imaging, coil sensitivity information can be obtained from calibration data for reconstruction that inevitably prolongs MRI scan. In recent years, structured low-rank matrix completion methods implicitly exploit coil sensitivity that enables calibrationless k-space estimation while prohibitively increases the computational burden. This study presents a fast and calibrationless image-space alternative for reconstruction that derives high-quality coil sensitivity and spatial support maps by structured low-rank tensor estimation. The proposed approach was evaluated with multi-channel multi-contrast brain datasets. It achieves a high convergence rate with significantly reduced reconstruction time, making the calibrationless reconstruction approach more efficient in clinical practice.
|
|
 |
0068.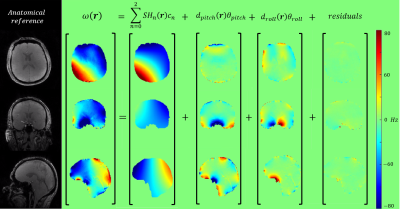 |
Data-driven motion-corrected brain MRI incorporating pose dependent B0 fields
Yannick Brackenier1,2, Lucilio Cordero-Grande1,2,3, Raphael Tomi-Tricot1,2,4, Tom Wilkinson1,2, Jan Sedlacik1,2, Philippa Bridgen1,2, Sharon Giles1,2, Shaihan Malik1,2, Enrico De Vita1,2, and Joseph V Hajnal1,2
1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 3Biomedical Image Technologies, ETSI Telecomunicación, Universidad Politécnica de Madrid and CIBER-BNN, Madrid, Spain, 4MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
A fully data-driven retrospective motion correction reconstruction for volumetric brain MRI at 7T that includes modelling of pose-dependent changes in polarising magnetic (B0) fields in the head has been developed. Building on the DISORDER framework, the use of a physics-based B0 model constrains the number of unknowns to be found, enabling motion correction based solely on data-consistency without requiring any additional probe- or navigator-data. The proposed reconstruction was validated on an in-vivo spoiled gradient echo acquisition in which the subject deliberately moved. Substantial removal of motion artefacts was achieved.
|
|
 |
0069.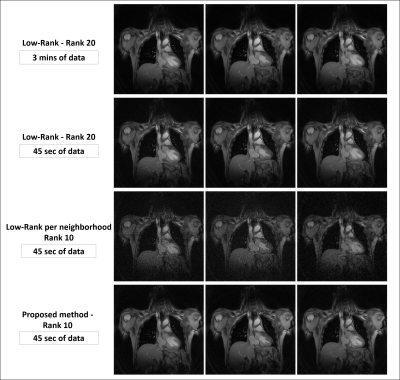 |
Manifold learning via tangent space alignment for accelerated dynamic MR imaging with highly undersampled (k,t)-data
Yanis Djebra1,2, Isabelle Bloch2,3, Georges El Fakhri1, and Chao Ma1
1Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States, 2LTCI, Telecom Paris, Institut Polytechnique de Paris, Paris, France, 3LIP6, Sorbonne University, CNRS, Paris, France
Many unsupervised learning methods have been proposed to discover the structure of manifolds embedded in high-dimensional input spaces. However, image reconstruction requires mapping the learned low-dimension data in the feature space back to the input space, which can be challenging if the mapping function is implicit. This work presents an image reconstruction scheme closely related to machine learning methods learning manifolds via tangent space alignment. Here, the mapping transform is explicit and learned from the data. This model is a nonlinear generalization of the Low-Rank matrix/tensor model, reconstructing undersampled MR data with lower rank than the standard Low-Rank reconstruction.
|
|
 |
0070.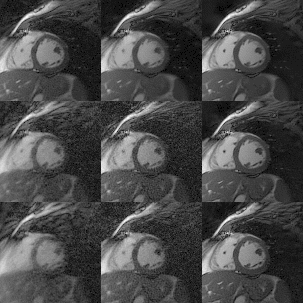 |
One-heartbeat cardiac CINE imaging via jointly regularized non-rigid motion corrected reconstruction
Gastao Cruz1, Kerstin Hammernik2,3, Thomas Kuestner1, Daniel Rueckert2,3, René M. Botnar1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Technical University of Munich, Munich, Germany, 3Department of Computing, Imperial College London, London, United Kingdom
Motion-resolved reconstructions are standard for cardiac CINE imaging by splitting data into multiple cardiac phases across several heartbeats. Consequently, the reconstruction of each phase is highly ill-posed, since only a subset of the data are used to reconstruct it. In this work a novel motion corrected framework is developed for highly accelerated cardiac CINE imaging. Each cardiac phase is reconstructed with a motion corrected reconstruction and jointly regularized with every other motion corrected cardiac phase via a non-rigidly aligned patch-based denoiser. This approach leads to a substantial improvement in image quality, enabling highly accelerated CINE images from a single heartbeat.
|
|
 |
0071.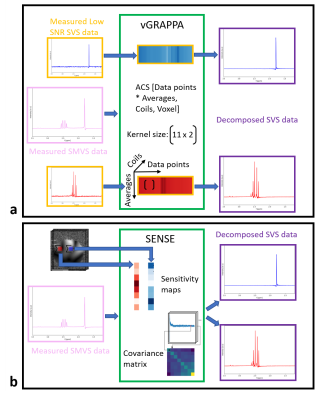 |
Fourier-based decomposition approach for simultaneous acquisition of 1H spectra from two voxels in vivo at short echo times
Layla Tabea Riemann1, Christoph Stefan Aigner1, Ralf Mekle2, Sebastian Schmitter1,3, Bernd Ittermann1, and Ariane Fillmer1
1Physikalisch-Technische Bundesanstalt (PTB), Braunschweig und Berlin, Germany, 2Center for Stroke Research Berlin, Charité Universitätsmedizin, Berlin, Germany, 3Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States
In this work, a Fourier-based technique for 1H MR spectroscopy based on split-slice-GRAPPA is introduced to decompose simultaneously acquired dual-voxel data. In contrast to the existing sensitivity-map-based approach, this technique does not need any additional image acquisitions. The autocalibration lines are derived by additional low SNR spectral data. 2SPECIAL was used to simultaneously acquire data from two voxels at short echo times. The proposed decomposition algorithm was first validated in a multi-compartment phantom, and its application was then demonstrated at 7T in vivo.
|
|
 |
0072.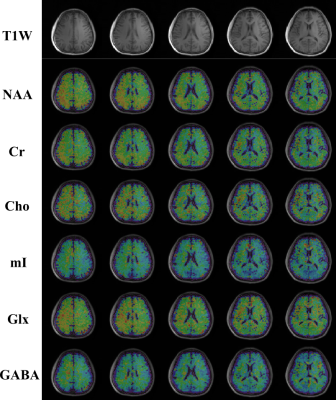 |
Optimized Subspace-Based J-Resolved MRSI for Simultaneous Metabolite and Neurotransmitter Mapping
Zepeng Wang1,2, Yahang Li1,2, and Fan Lam1,2
1Department of Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, Urbana, IL, United States
J-resolved 1H-MRSI offers improved molecular specificity by encoding the J-coupling evolution of different molecules at multiple TEs. The addition of another encoding dimension poses both challenges and flexibility for optimizations in data acquisition and reconstruction. This work presents further optimized J-resolved MRSI acquisition and reconstruction strategies for high-resolution, 3D metabolite, and neurotransmitter mapping. Specifically, estimation-theoretic TE selection within a union-of-subspaces (UoSS) framework was analyzed for optimized separation of metabolite and neurotransmitter signals. Both simulation and in vivo studies have been conducted. Promising results in terms of simultaneously high-resolution mapping of major metabolites, Glx, and GABA are provided.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.